Family: Phasmaviridae
Genus: Feravirus
Distinguishing features
Ferak virus (FRKV), Guagua virus (GUAV), hemipteran phasma-related virus OKIAV247 (HeFV), and neuropteran phasma-related virus OKIAV248 (NeFV) are the only classified feraviruses. These viruses are associated with dipteran, hemipteran, and neuropteran insects (Marklewitz et al., 2015, Käfer et al., 2019).
Virion
Feravirions are spherical or pleomorphic in shape (60–120 nm in diameter) and surrounded by a membrane envelope that is decorated with glycoprotein (GP) spikes composed of subunits GN and GC (Figure 1 Feravirus) (Marklewitz et al., 2015).
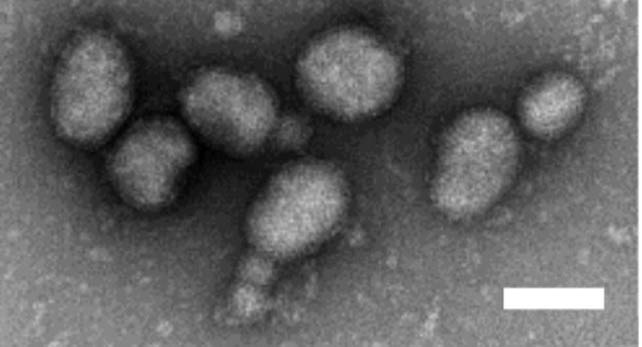 |
| Figure 1 Feravirus. Electron micrograph of negative-stained virions of Ferak virus sedimented by ultracentrifugation. Scale bar: 100 nm (image reproduced from 26038576). |
Nucleic acid
Feraviruses have tri-segmented negative-sense RNA genomes of about 12.3–12.7 kb (small [S] segment: 1.5–1.6 kb; medium [M] segment: 4.2–4.3 kb; large [L] segment: 6.6–7.0 kb) (Käfer et al., 2019).
Proteins
Feraviruses express three structural proteins: nucleoprotein (N), glycoprotein precursor (GPC), and L protein; and two nonstructural proteins, NSs and NSm (Marklewitz et al., 2015).
Carbohydrates
GN and GC envelope proteins are predicted to be glycosylated, but these proteins are resistant to N-glycosidase treatment (Marklewitz et al., 2015).
Genome organization and replication
The S segment encodes N and NSs, the M segment encodes GPC and NSm, and the L segment encodes L protein (Käfer et al., 2019) (Figure 2 Feravirus). Feravirus genomic segments are expected, by analogy with other bunyaviricetes, to assume circular ribonucleoprotein complexes within the virion via non-covalent binding of complementary and conserved 3′- and 5′-terminal sequences. Complementary terminal sequences for the three segments of FRKV are 5′-AGUAGUAAACA (Marklewitz et al., 2015). During FRKV replication in C6/36 mosquito cell culture, viral RNA and mRNA were shown to be similar in length, and mRNAs included 6–23 5′ non-viral nucleotides obtained by cap-snatching. FRKV replication was blocked at temperatures above 30˚C, indicating an insect-only transmission cycle (Marklewitz et al., 2015).
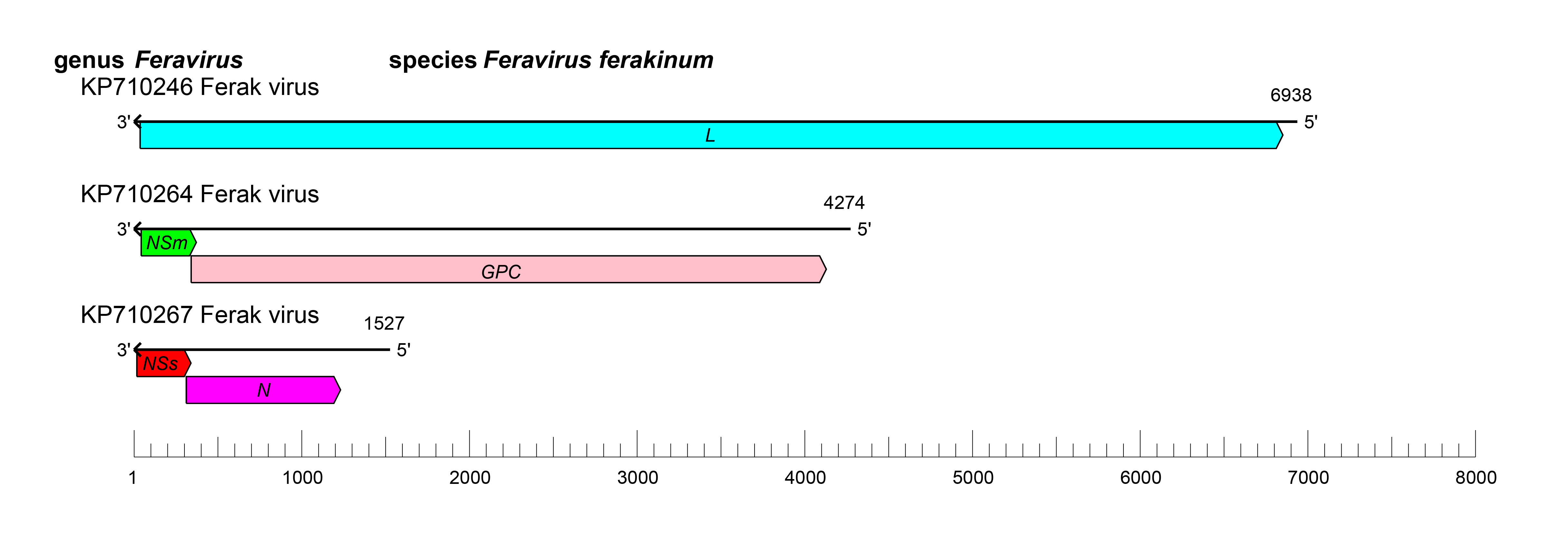 |
| Figure 2 Feravirus. Schematic representation of feravirus genome organization. L: gene encoding L protein; GPC, gene encoding GPC; N: gene encoding N protein; NSm: gene encoding NSm; NSs: gene encoding NSs. |
Biology
FRKV has been detected in culicid mosquitoes (Anopheles gambiae Giles 1902, Culex spp., Uranotaenia mashonaensis Theobald, 1901) in Côte d’Ivoire (Marklewitz et al., 2015). GUAV has been detected in culicid mosquitoes (Mansonia titillans Walker, 1848) in Columbia [unpublished]. HeFV has been detected in hawthorn shield bugs (acanthosomatid Acanthosoma haemorrhoidale (Linnaeus, 1758)) in Germany (Käfer et al., 2019). NeFV has been detected in green lacewings (Peyerimhoffina gracilis (Schneider, 1851)) in Austria (Käfer et al., 2019).
Species demarcation criteria
The feravirus species demarcation criterion is <95% identity in the amino acid sequence of the L protein.

