Family: Peribunyaviridae
Genus: Pacuvirus
Distinguishing features
Pacuvirus genomes encode four structural proteins: the RNA-directed RNA polymerase/endonuclease L, the glycoproteins Gn and Gc, and the nucleoprotein N (Table 2 Peribunyaviridae; Figure 2 Peribunyaviridae). Viruses in the genus encode putative non-structural (NS) NSm proteins, but not NSs protein (Rodrigues et al., 2014).
Virion
Unknown.
Genome organization and replication
The pacuvirus genome is similar to that of other viruses in the family. The L segment encodes the L protein, the M segment encodes the glycoproteins Gn and Gc and the non-structural protein NSm, and the S segment encodes N; the genome does not encode an NSs protein (Table 2 Peribunyaviridae; Figure 1 Pacuvirus) (Rodrigues et al., 2014).
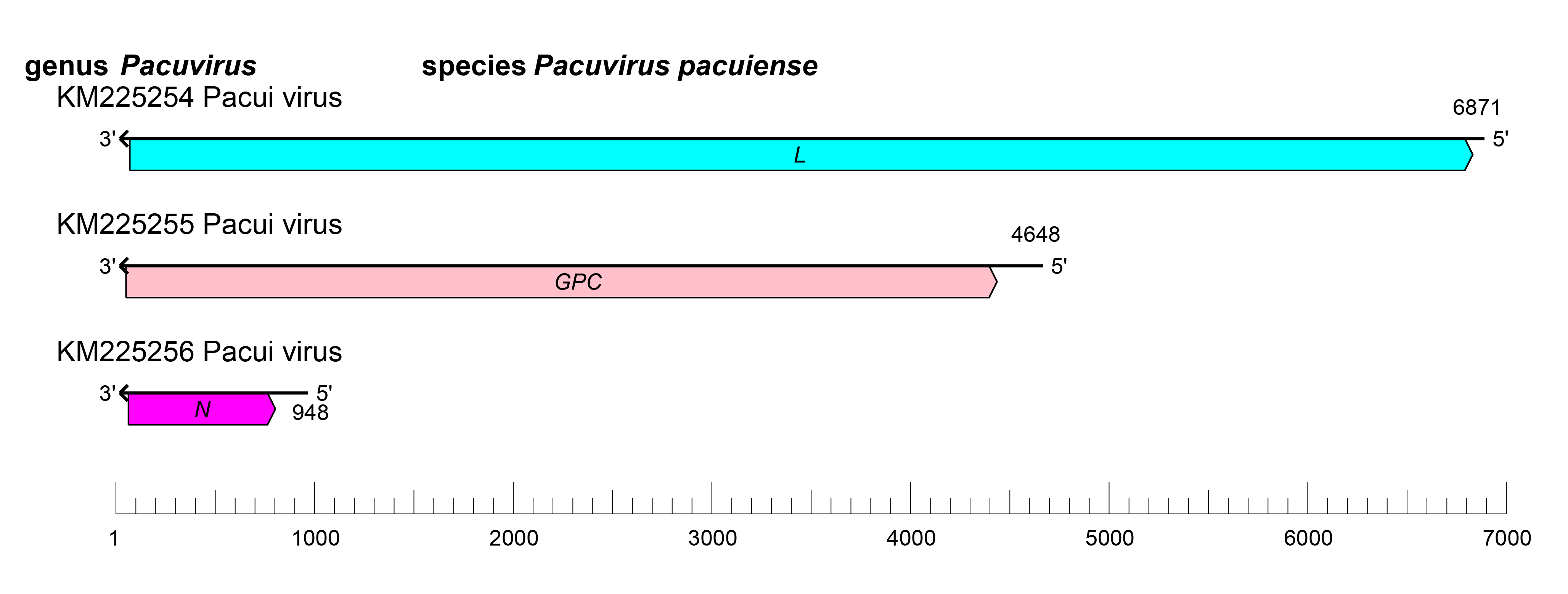 |
| Figure 1 Pacuvirus. Pacuvirus coding strategy. vcRNAs are depicted in 3′→5′ direction and mRNAs are depicted in a 5′→3′ direction. Coloured boxes on the mRNAs depict ORFs that encode the N, nucleocapsid protein; Gn and Gc, external glycoproteins; L, large protein, as well as the non-structural protein NSm. |
Replication and morphogenesis in the Golgi are likely to be similar to that of other members of the family; however, this has not been thoroughly examined (see ‘Genome organization and replication’ on the family page).
Biology
Pacuviruses have been isolated from psychodid sandflies and from rodents in Brazil (Aitken et al., 1975, Rodrigues et al., 2014). Serological surveys have found antibodies to Pacui virus in rodents and marsupials, but not in humans (Aitken et al., 1975). Pacui virus was originally assigned to the genus Phlebovirus (family Phenuiviridae) due to the association with sandflies; however, recent sequence analyses have revealed that pacuviruses have genomes organized similar to that of the orthobunyaviruses (Figure 2.Peribunyaviridae; (Rodrigues et al., 2014).
Antigenicity
The glycoproteins and nucleocapsid protein can elicit an antibody response (Aitken et al., 1975). Where studied, pacuviruses do not react by complement fixation with many orthobunyaviruses (Tesh et al., 1975, Travassos da Rosa et al., 1983).
Species demarcation criteria
Species are defined by pairwise evolutionary distance values of greater than 0.1 when using the WAG substitution model on concatenated amino acid sequences of all three segments (Maes and Kuhn 2018).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Santarem virus | L: MK896471; M: MK896470; S: MK896469 | STMV |
Virus names and virus abbreviations are not official ICTV designations.

