Family: Peribunyaviridae
Genus: Orthobunyavirus
Distinguishing features
Orthobunyaviruses have a wide geographic and host range, although individual viruses may be restricted to a small number of host species. Viruses in the genus form a monophyletic clade upon comparison of L protein amino acid sequences, and encode an NSm, and often an NSs protein. Segment reassortment is well-described among related strains and has been associated with disease emergence.
Virion
Morphology
Orthobunyavirions are spherical or pleomorphic in shape and 80−120 nm in diameter (Martin et al., 1985). The enveloped surface (about 5 nm thick) has projections of glycoprotein heterodimers (Gn and Gc) about 18 nm long with an estimated 650 copies per virion (Talmon et al., 1987). The surface is a closely packed lattice of glycoprotein Gn-Gc spikes organized in a tripod-like order (Bowden et al., 2013). Structural analysis has revealed that the N-terminal variable half of the Gc glycoprotein is composed of two domains: a 27 kDa α-helical head domain and a stalk that is divided into two identically folded 19 kDa tandem β-sandwiches (Hellert et al., 2019). The projecting spike is also the major target of the neutralizing antibody response (Hellert et al., 2019).
Nucleic acid
Orthobunyaviruses have the peribunyavirus-typical tripartite, negative-sense genome organization. The terminal nucleotides of each segment (S, M, and L) occur in a canonical, conserved sequence (in coding sense) 5′-AGTAGTGT…ACACTACT-3′. As with other peribunyaviruses, genomic vRNAs are not polyadenylated nor are they modified at the 5′-end. Viral mRNAs are not polyadenylated, are truncated relative to the vRNA (Abraham and Pattnaik 1983, Eshita et al., 1985). mRNAs possess a 5′-methylated cap derived from host mRNA via “cap snatching”, which is mediated by the endonuclease function of the L protein (Reguera et al., 2010).
Proteins
Like other peribunyaviruses, all orthobunyavirions contain of four structural proteins: Gn, Gc, nucleocapsid (N), and RNA-directed RNA polymerase/endonuclease protein (L) (Figure 1 Peribunyaviridae, Figure 2 Peribunyaviridae; Table 2 Peribunyaviridae). Non-structural proteins (NSs and NSm) are encoded in the S and the M segments of some orthobunyaviruses (Figure 1 Orthobunyavirus; Table 2 Peribunyaviridae).
Lipids
Orthobunyavirion lipids are derived from the host membranes where viruses mature, and include phospholipids, sterols, fatty acids, and glycolipids. Virions contain 20–30% lipids by weight (Bishop et al., 1980).
Carbohydrates
Orthobunyavirions may contain asparagine-linked mannose sugars on the Gn and Gc proteins when grown in vertebrate cells. Carbohydrates make up 7% of the virion weight (Bishop et al., 1980).
Genome organization and replication
The S segment encodes N, and may encode the non-structural protein NSs in an overlapping ORF downstream (Fuller et al., 1983) or upstream of the N protein initiation codon (Aguilar et al., 2018) (Figure 1.Orthobunyavirus). However, not all orthobunyaviruses encode NSs (Mohamed et al., 2009, de Melo et al., 2018). The M segment encodes Gn and Gc proteins (Fazakerley et al., 1988) (Figure 1 Orthobunyavirus), that are co-translationally cleaved by host signalase (Fazakerley and Ross 1989) and signal peptide peptidase (Shi et al., 2016). Orthobunyaviruses also encode an NSm protein between the Gn and Gc coding regions (Figure 1 Orthobunyavirus). The L segment encodes the L protein (Figure 1 Orthobunyavirus).
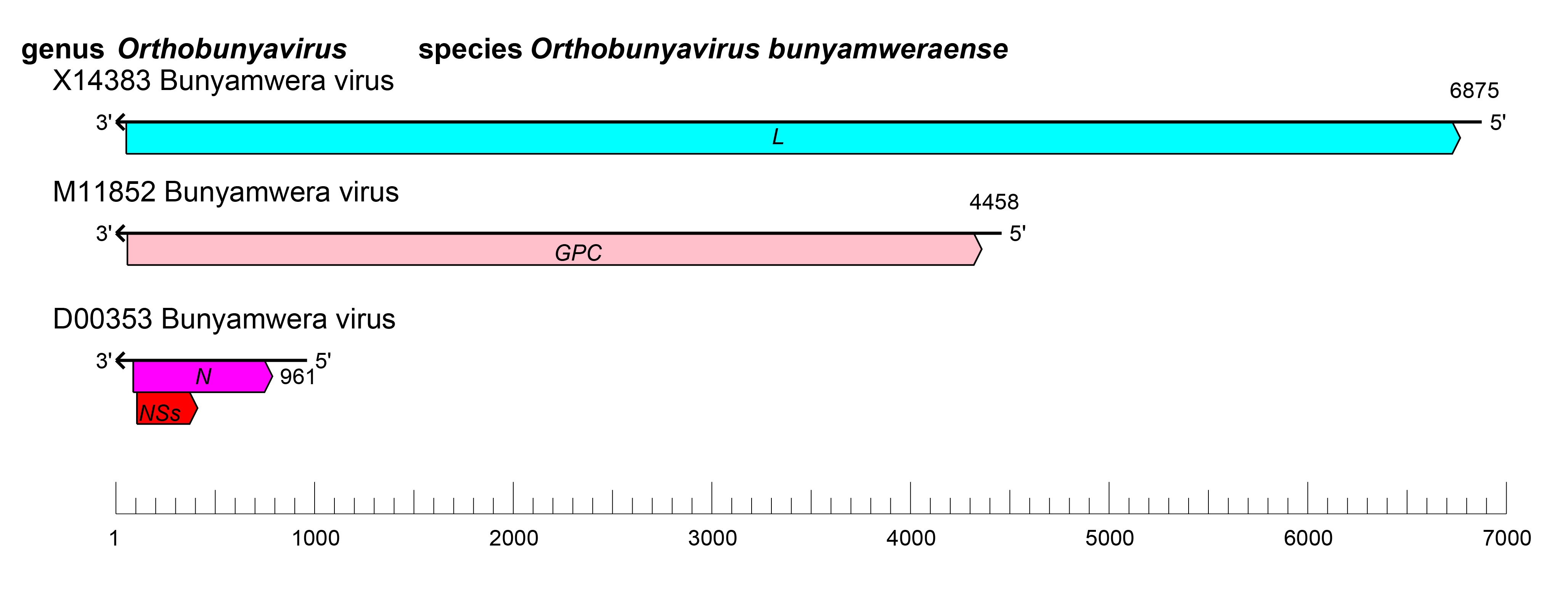 |
| Figure 1 Orthobunyavirus. Orthobunyavirus coding strategy. vcRNAs are depicted in 3′→5′ direction and mRNAs are depicted in a 5′→3′ direction. Coloured boxes on the mRNAs depict ORFs that encode the N, nucleocapsid protein; Gn and Gc, external glycoproteins; L, large protein, as well as the non-structural proteins NSs and NSm. |
Replication, morphogenesis, assembly, and budding are described in ‘Genome organisation and replication’ on the family page).
Biology
The genus Orthobunyavirus is the largest and most diverse in the family including 138 species, members of which occur globally in tropical, temperate and arctic ecological niches. Although a wide range of arthropod and vertebrate hosts are described for viruses of the genus, each virus possesses a restricted arthropod and host range (Beaty and Calisher 1991), which in turn limits its geographic distribution (Calisher 1996). However, some viruses share common hosts and overlapping geographies. Orthobunyaviruses have been isolated from many different vertebrate hosts including squirrels (La Crosse virus), bats (Mojuí dos Campos virus), rabbits (snowshoe hare virus), ungulates (Akabane virus), sloths (Oropouche virus), and birds (Mermet virus). Most orthobunyaviruses are transmitted by mosquitoes. However, biting midges, bed bugs, and wingless bat flies are also known vectors. Some strains are known to be maintained in the arthropod host through transovarial transmission (Watts et al., 1974). Orthobunyaviruses are associated with a broad spectrum of human disease, including encephalitides (e.g., La Crosse virus), febrile illnesses (e.g., Bunyamwera virus), and viral haemorrhagic fever (e.g., Ngari virus Garissa variant). Adverse veterinary outcomes include fetal abnormalities and abortion storms among livestock (e.g., Cache Valley and Schmallenberg viruses). Coinfection with more than one orthobunyavirus can lead to reassortment of genomic segments in both laboratory and natural settings (Beaty et al., 1981, Beaty et al., 1985, Bishop and Beaty 1988, Borucki et al., 1999, Cheng et al., 1999, Nunes et al., 2005, Briese et al., 2007, Yanase et al., 2012). Mosquitoes are reservoirs for the reassortment of genomic segments between heterotypic orthobunyaviruses of shared serological character (Beaty et al., 1981, Beaty et al., 1985, Borucki et al., 1999). In addition, orthobunyavirus segment reassortment has been associated with outbreaks of human disease (Gerrard et al., 2004, Briese et al., 2006).
Antigenicity
The Gn and Gc proteins are responsible for eliciting hemagglutination-inhibiting and neutralizing antibodies (Kingsford et al., 1983). Complement-fixing antibodies are elicited by the nucleocapsid protein. Orthobunyaviruses can be placed into one of at least 18 serogroups, or remain ungrouped, by the use of hemagglutination-inhibition, neutralization and complement fixation assays (Calisher 1996).
Species demarcation criteria
Phylogenetic analysis of viruses in the family Peribunyaviridae is shown in Figure 2 Orthobunyavirus.
Species within the genus Orthobunyavirus can be defined by:
- Less than 96% identity in the complete amino acid sequence of the L protein
The species Orthobunyavirus acaraense, Orthobunyavirus cuchillaense, Orthobunyavirus bakauense, Orthobunyavirus batamaense, Orthobunyavirus bertiogaense, Orthobunyavirus minatitlanense, Orthobunyavirus mpokoense, Orthobunyavirus olifantsvleiense and Orthobunyavirus turlockense were originally established according to previous serological demarcation criteria and have not been evaluated using genomic speciation demarcation criterion.
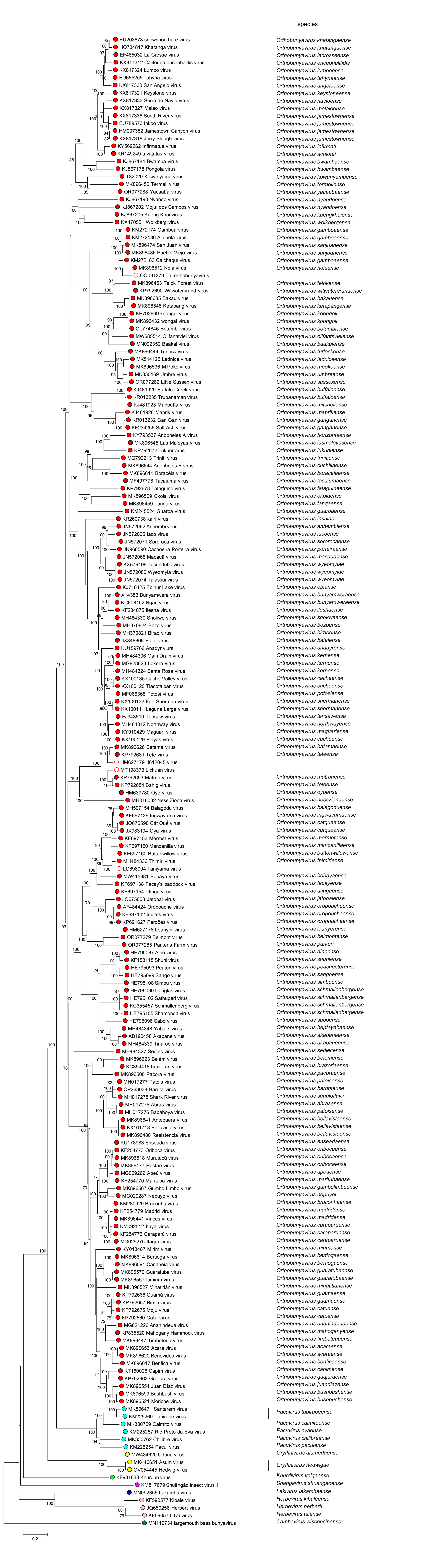 |
| Figure 2 Orthobunyavirus. Phylogenetic analysis of members of the family Peribunyaviridae. The amino acid sequences comprising the L protein of family members were aligned with Clustal W and a neighbor joining tree was produced based on the JTT model using Mega version 7. Numbers at nodes indicate bootstrap support where this was > 70%. Members of the same genus have circles with the same colour: open circles indicate unclassified viruses. |
Related, unclassified viruses
| Virus name | Accession number | Virus Abbreviation | Reference |
| Belmont virus | L: KT720482; M: KT720481* | BELV | (Huang 2016) |
| Kedah fatal kidney syndrome virus | L: MK047412; M: MK047406; S: MK047401* | KFKSV | (Palya et al., 2019) |
| little Sussex virus | L: KT720483* | LTLSV | (Huang 2016) |
| Ntwetwe virus | L: MH324826; M: MH324827; S: MH324828* | NTWV | (Edridge et al., 2019) |
| Nyangole virus | L: MH685711; M: MH685710; S: MH685709*
| NYGV | (Ramesh et al., 2019) |
| Parker's Farm virus | L: KT720484* | PFV | (Huang 2016) |
| Wǔhàn louse fly virus 1 | L: KM817663; M: KM817725; S: KM817755* | WLFV | (Li et al., 2015) |
| Yacaaba virus | L: KT820208; M: KT820209; S: KT820210* | YACV | (Huang et al., 2016) |
* partial genome
Virus names and virus abbreviations are not official ICTV designations.

