Family: Partitiviridae
Genus: Betapartitivirus
Distinguishing features
Members of the genus Betapartitivirus infect plants and fungi. Betapartitiviruses have two dsRNA segments that are believed to be individually encapsidated in separate particles, and genomes typically ranging from 4.3–4.8 kbp in total.
Virion
Morphology
Based on negative-contrast electron microscopy, the virions of white clover cryptic virus 2 are isometric and approximately 38 nm in diameter (Boccardo et al., 1985). The particles appear rounded and are not penetrated by stain (Figure 1 Betapartitivirus). The structure of Fusarium poae virus 1 has been determined by electron cryomicroscopy and 3D reconstruction (Tang et al., 2010). The outer diameter of the capsid is about 40 nm, and each particle contains 120 CP molecules arranged with icosahedral symmetry (Tang et al., 2010). Other reported betapartitivirus particle sizes range from 25 to 35 nm in diameter (Strauss et al., 2000, Lim et al., 2005, Sasaki et al., 2005, Li et al., 2009, Ziegler et al., 2012).
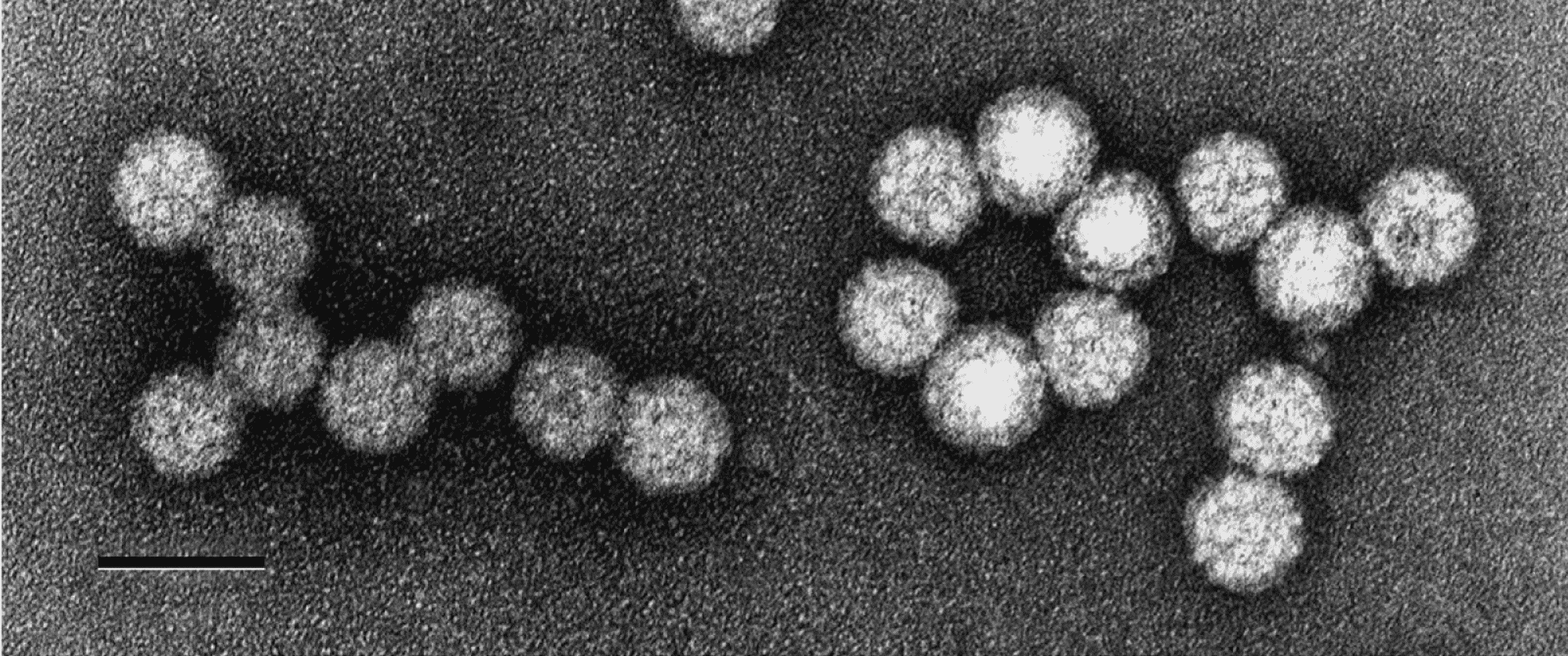 |
| Figure 1 Betapartitivirus. Negative-contrast electron micrograph of particles of an isolate of Betapartitivirus trifolii. The bar represents 50 nm. (Ghabrial et al., 2000). |
Physicochemical and physical properties
Virion buoyant density of white clover cryptic virus 2 in CsCl is 1.375 g cm−3 (Boccardo et al., 1985).
Nucleic acid
Viral genome comprises two dsRNA segments, which are each about 2.1–2.4 kbp.
Proteins
There is a single major coat protein (CP) with predicted Mr of 71–77 kDa. The predicted Mr of the RNA-dependent RNA polymerase (RdRP), as deduced from nucleotide sequence analysis, ranges from 77 to 87 kDa. Virion-associated RNA polymerase activity is present.
Genome organization and replication
Each genome segment is monocistronic: the larger genome segment typically encodes the RdRP and the smaller one encodes the putative CP. Atkinsonella hypoxylon virus has a third dsRNA segment appearing not to encode any proteins (see GenBank record L39127) (Oh and Hillman 1995). Members of genus Betapartitivirus commonly have poly(A) tracts (≥10 nt) near to the coding-strand 3′-terminus of one or both genome segments.
Biology
Members of the genus Betapartitivirus infect ascomycetous and basidiomycetous fungi (ten species) and plants (seven species). They are transmitted intracellularly during cell division (plants, fungi) or cell fusion and sporogenesis (fungi). Plant betapartitiviruses include former members of the defunct genus Betacryptovirus, which are associated with persistent infections (Lesker et al., 2013). Various host effects have been described for fungal betapartitiviruses: Rosellinia necatrix virus 1 mediates changes in host colony morphology and decreased dispersal via conidiospores (Kanematsu et al., 2010), and the related, unclassified Sclerotinia sclerotiorum partitivirus 1 reduces the virulence of its host (Xiao et al., 2014).
Species demarcation criteria
The species demarcation criteria within genus Betapartitivirus are:
- ≤ 90% aa sequence identity in the RdRP, and
- ≤ 80% aa sequence identity in the CP
Related, unclassified viruses
Virus name* | Accession number | |
| dsRNA1/RdRP | dsRNA2/CP | |
| Sclerotinia sclerotiorum partitivirus 1 strain SsPV1-WF-1 | JX297511 | JX297510 |
| Ustilaginoidea virens partitivirus 3 strain HP-30 | KF680478 | KF680479 |
| Rosellinia necatrix partititivirus 6 strain W113 | LC010952‡ | LC010953 |
| Fusarium poae partitivirus 2 | LC150608 | LC150609 |
Virus names and virus abbreviations are not official ICTV designations.
* Representative isolate of a virus with a complete published genome sequence
‡ Defective-interfering RNAs derived from dsRNA1 deposited under GenBank accessions LC062720-27

