Family: Partitiviridae
Genus: Gammapartitivirus
Distinguishing features
All known members of the genus Gammapartitivirus infect ascomycetous fungi. Gammapartitiviruses have two dsRNA segments that are believed to be individually encapsidated in separate particles. The genome is typically 3.1–3.4 kbp in total.
Virion
Morphology
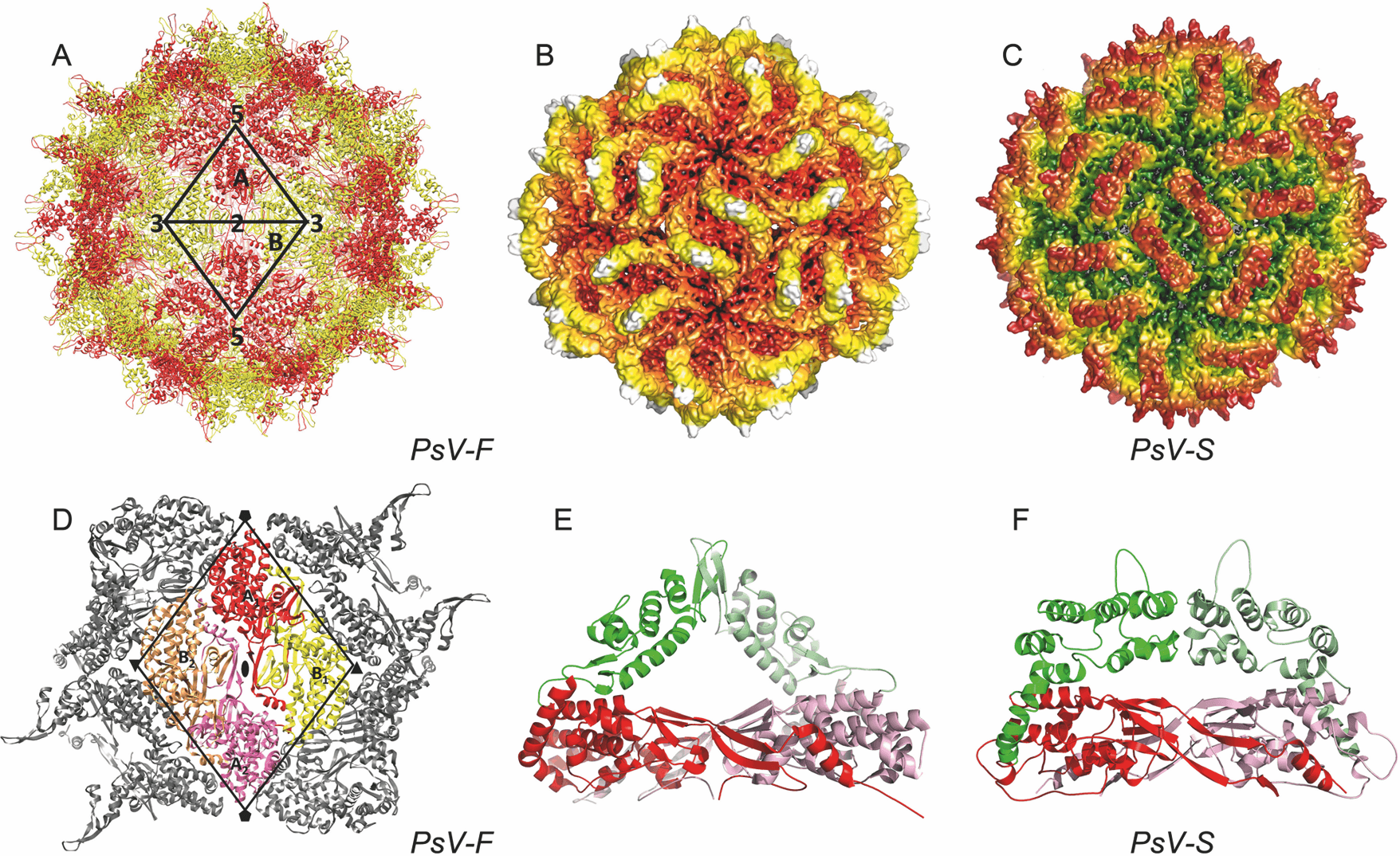
The structures of Penicillium stoloniferum virus F (PsV-F) and Penicillium stoloniferum virus S (PsV-S) have been determined by X-ray crystallography (PsV-F) and electron cryomicroscopy (PsV-F and PsV-S) (Ochoa et al., 2008, Pan et al., 2009). The outer diameter of the capsid is about 35–40 nm. The most prominent features of the capsid are 60 arch-like protrusions that decorate a spherical shell (Figure 1 Gammapartitivirus panels A–C). Each particle contains 120 coat protein (CP) molecules arranged with icosahedral symmetry. The tertiary structure of each CP molecule consists of two distinct domains, one of which forms the continuous, 3 nm thick capsid, and the other that, with the corresponding domain of a neighboring CP, comprises the arch (Figure 1 Gammapartitivirus panels E–F). The asymmetric unit of the icosahedron consists of two related CP molecules, CPA and CPB. The 60 CPA molecules are organized as flower-like pentamers, each centered about one of the 12 vertices of the icosahedral capsid (Figure 1 Gammapartitivirus panel A). The 60 CPB molecules are arranged as trimers at the twenty icosahedral 3-fold (3f) axes of symmetry (Figure 1 Gammapartitivirus panel A). The CPA-CPB dimer, defined by two monomers that form an arch, is most stable based on buried surface areas and likely functions as the assembly precursor (Figure 1 Gammapartitivirus panels E–F). The two CP molecules in this dimer assume nearly identical conformations and are related by almost-perfect local 2f symmetry. Each dimer is stabilized by extensive structure swapping between the monomers and the additional intersubunit interactions mediated by the arch tip. Unlike the assembly pathway that has been described for reoviruses and other larger dsRNA viruses, PsV-F and PsV-S assembly is likely to proceed from dimers of CP dimers, each of which adopts a striking, smooth-edged diamond shape (Figure 1 Gammapartitivirus panel D). The N-terminal region (approximately 40 aa) of the CP polypeptide extends radially inwards and interacts with the dsRNA viral genome. Given the large number of basic residues within this region, it may participate in RNA packaging during particle assembly, or play a role during viral transcription. Small pores in the capsid shell at the icosahedral 5f and 3f axes may facilitate the export of RNA transcripts. The dsRNA genome appears as concentric layers in icosahedrally averaged maps.
Physicochemical and physical properties
Penicillium stoloniferum virus S virions with double-stranded RNA have a buoyant density of ~1.36 g cm-3 in CsCl (Buck and Kempson-Jones 1973)
Nucleic acid
Viral genome comprises two dsRNA segments, which are 1.6–1.8 (dsRNA1) and 1.4–1.6 kbp (dsRNA2).
Proteins
There is a single major CP with predicted Mr ranging from 44 to 47 kDa. The Mr of the RNA-dependent RNA polymerase (RdRP), as predicted from nucleotide sequence analysis, ranges from 60 to 62 kDa. Virion-associated RNA polymerase activity is present.
Genome organization and replication
Each genome segment is monocistronic: the larger one typically encodes the RdRP and the smaller one the putative CP. Apparent RNA satellites encoding small (237–303 aa) proteins, homologous to each other but not to either CP or RdRP, are present in classified gammapartitiviruses and related, unclassified gammapartitiviruses including Aspergillus ochraceous virus (EU118279), Discula destructiva virus 1 (AF316994), Gremmeniella abietina RNA virus MS1 (AY089995) and Ustilaginoidea virens partitivirus 1 (KC503901). Moreover, RNA satellites appearing not to encode any proteins are found associated with studied isolates of Discula destructiva virus 1 and Penicillium stoloniferum virus F (AF316995 and AY738338, respectively). Gammapartitiviruses do not usually have poly(A) tracts near the plus-strand 3′-terminus of their genome segments.
Biology
Members of the genus Gammapartitivirus infect ascomycetous fungi. They are transmitted intracellularly during cell division or cell fusion. The related, unclassified Aspergillus fumigatus partitivirus 1 seems to mediate reduced growth rate, conidiation and pigmentation in Aspergilllus fumigatus (Bhatti et al., 2011).
Species demarcation criteria
The species demarcation criteria within genus Gammapartitivirus are:
- ≤ 90% aa sequence identity in the RdRP, and
- ≤ 80% aa sequence identity in the CP
Related, unclassified viruses
| Virus name* | Accession number | |||
| dsRNA1/RdRP | dsRNA2/CP | dsRNA3 | dsRNA4 | |
| Ustilaginoidea virens partitivirus 1 | KC503898 | KC503899 | KC503900‡ | KC503901‡ |
| Aspergillus fumigatus partitivirus 1 isolate 88 | FN376847 | FN398100 | ||
| Colletotrichum acutatum partitivirus 1 isolate CaRV1 | KC572132 | KC572133 | ||
| Ustilaginoidea virens partitivirus 2 isolate Uv0901 | KF361014 | KF361015 | ||
| Verticillium dahliae partitivirus 1 strain Vd08284 | KC422244 | KC422243 | ||
| Penicillium aurantiogriseum partitivirus 1 | KT601103 | KT601104 | ||
Virus names and virus abbreviations are not official ICTV designations.
* Representative isolate of a virus with a complete published genome sequence
‡ Segment encoding a hypothetical protein with unknown function