Family: Nodaviridae
Genus: Betanodavirus
Distinguishing features
Betanodaviruses are pathogens of fish causing a disease called “viral nervous necrosis” and are named after the host fish from which they were isolated, followed by nervous necrosis virus (Munday et al., 2002). They infect more than 50 species of marine and freshwater fish. Five genetic lineages of betanodavirus have been identified.
Virion
Morphology
Virions are non-enveloped, spherical in shape, 25–33 nm in diameter, and have icosahedral symmetry (T=3). Distinct surface protrusions are observed by electron microscopy of negatively stained preparations (Figure 1. Betanodavirus). Image reconstruction of virus-like particles of Malabaricus grouper nervous necrosis virus (MGNNV) indicates that the capsid protein (CP) of betanodaviruses has a two domain structure compared to the single domain structure of the CP of alphanodaviruses. The average diameter of the particle is 31 nm. In contrast with most alphanodaviruses, empty particles have been seen by electron microscopy of some preparations of betanodaviruses.
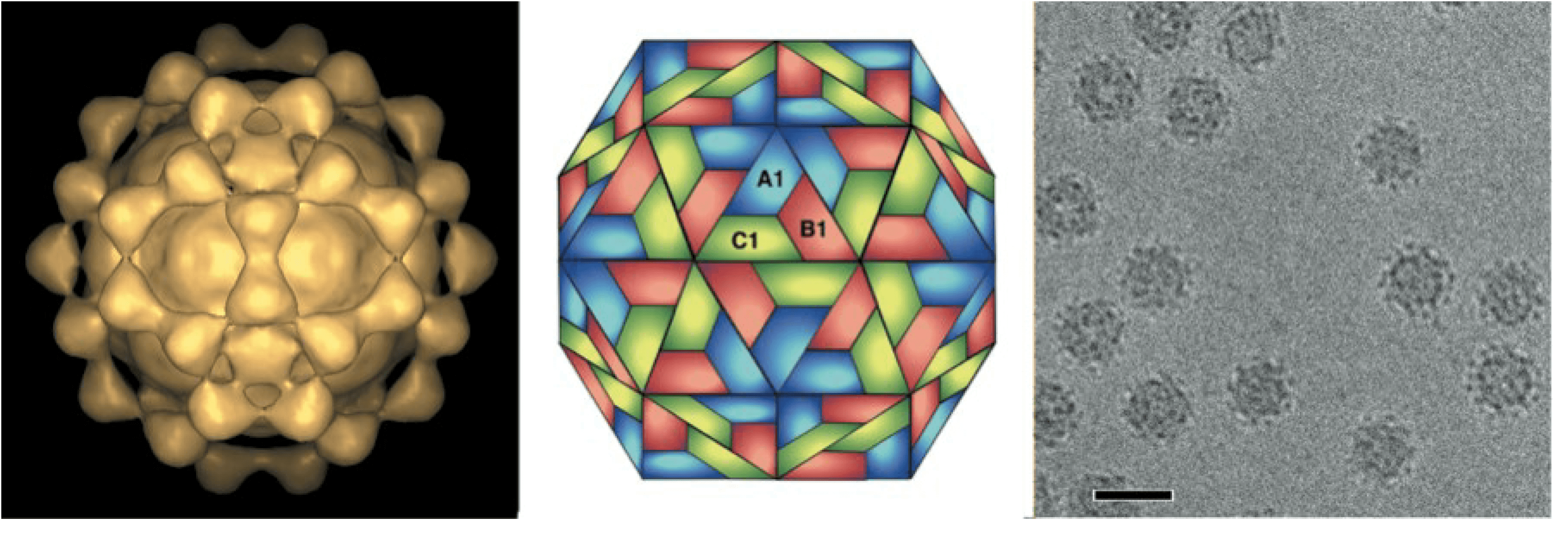 |
| Figure 1. . Betanodavirus Malabaricus grouper nervous necrosis virus particles. (Left) Image reconstruction of virus-like particles generated in Spodoptera frugiperda cells from a recombinant baculovirus expressing the Malabaricus grouper nervous necrosis virus coat protein gene. (Center) Schematic representation of a T=3 icosahedral lattice. A1, B1 and C1 indicate three different quasi-equivalent copies of the capsid protein. (Right) Cryo-electron micrograph of virus-like particles; the bar represents 40 nm. (Courtesy of L. Tang and J.E. Johnson) |
Physicochemical and physical properties
Virion buoyant density in CsCl of striped jack nervous necrosis virus (SJNNV) has not been reported but that of Dicentrarchus labrax encephalitis virus (DlEV), a related but not yet classified betanodavirus, is about 1.31–1.36 g cm−3. Virions of DlEV are stable between pH 2 and 9 and resistant to heating at 56 °C for 30 min. Infectivity is resistant to extraction of virions with chloroform.
Nucleic acid
The genome consists of two molecules of positive-sense ssRNA: RNA1 (Mr 1.01×106, 3.1 kb) and RNA2 (Mr 0.49×106, 1.4 kb). Both RNA molecules are encapsidated in the same virus particle, and both are required for infectivity. Both molecules are capped at their 5′-ends and lack poly(A) tails at their 3′-ends.
Proteins
Betanodavirus capsids contain 180 copies of a single structural protein of 42 kDa. In contrast to alphanodaviruses, maturation cleavage of this protein is not observed.
Lipids
None.
Carbohydrates
None.
Genome organization and replication
Betanodaviruses replicate in the cytoplasm. Infected cells contain three ssRNAs: RNA1 (Mr 1.01×106; 3.1 kb); RNA2 (Mr 0.49×106; 1.4 kb) and a subgenomic (sg) RNA3 (Mr about 0.13×106; 0.4 kb) derived from RNA1. RNA3 is not packaged into virions. RNA1 encodes protein 1a (110 kDa), the RNA-dependent RNA polymerase (RdRP). RNA2 encodes protein 2a (42 kDa), the CP. The RNA3 of SJNNV encodes protein B2 and has a potent RNA silencing-suppression activity, as also observed for alphanodaviruses (Iwamoto et al., 2001).
Biology
Host range
Isolates of all species of betanodaviruses have been isolated from larvae, juvenile or adult marine fish, in which they cause “viral nervous necrosis” or “viral encephalopathy and retinopathy” associated with behavioral abnormalities and high mortalities. SJNNV and TPNNV have a limited host range: striped jack for SJNNV and tiger puffer for TPNNV. In contrast, RGNNV and BFNNV have a wide range of host fish; RGNNV is isolated from warm-water fishes and BFNNV is isolated from cold-water fishes. These diseases cause significant problems for the marine aquaculture industry.
Betanodaviruses replicate in cultured cells from striped snakehead fish (SNN-1 and E-11) and other cells derived from fish such as groupers (GF-1), sea bass (SBL), turbot (TV-1), and gilthead sea bream (SAF-1). A low level of virus replication is observed in mammalian (COS-1 and HeLa) cells.
Transmission
Betanodavirus antigens and/or CP genes are detected in eggs, larvae and ovaries of some inapparently infected hatchery-reared fish species, and the CP gene is frequently detected in a variety of wild fishes without any disease signs, indicating both horizontal and vertical modes of transmission of the virus.
Antigenicty
Betanodaviruses are cross-reactive by immunoblot analysis using polyclonal antisera but differential reactivity is observed with monoclonal antibodies. Virus neutralization with polyclonal antisera divides four betanodaviruses into three serotypes; serotype A for SJNNV, serotype B for tiger puffer nervous necrosis virus (TPNNV), and serotype C for red-spotted grouper nervous necrosis virus (RGNNV) and barfin flounder nervous necrosis virus (BFNNV).
Species demarcation criteria
The following criteria can be applied to the demarcation of species within the Betanodavirus genus:
- Biological properties (host range, vectors, mode of transmission). They primarily infect fish especially larval and young fish.
- Antigenic properties. Strains of betanodavirus fell into 3 major serotypes (A, B, C) and this sero-grouping is in part consistent with their genotypes, i.e. serotype A for striped jack nervous necrosis virus (SJNNV) genotype, serotype B for tiger puffer nervous necrosis virus (TPNNV) genotype, and serotype C for both red-spotted grouper nervous necrosis virus (RGNNV) and barfin flounder nervous necrosis virus (BFNNV) genotypes. The serological relatedness between RGNNV and BFNNV genotypes may result from their relatively higher similarity in RNA2 sequences.
- Virion electrophoretic mobility. Intact virus particles migrate with characteristic electrophoretic mobilities in non-denaturing agarose gel.
- Sedimentation coefficient, buoyant density. Virion sedimentation coefficient and buoyant density should be compared with those of other nodavirus species.
- Structural protein characteristics. The electrophoretic mobilities in SDS-PAGE of the CP precursor or its cleavage products should be compared with those of other nodavirus species.
- RNA electrophoretic mobilities. In the absence of sequence information, the electrophoretic mobilities of the viral genomic RNAs should be compared with those of other nodaviruses.
- Genome sequence. The nucleotide sequence of the two genomic RNAs should be compared with those of other nodaviruses (Thiéry et al., 2004) (Figure 2. Betanodavirus). Different species encode capsid proteins that differ at >15% of nucleotides and > 12% of amino acid positions. Because the nodavirus genome is segmented, reassortment can occur and the two genome segments may have distinct evolutionary lineages.
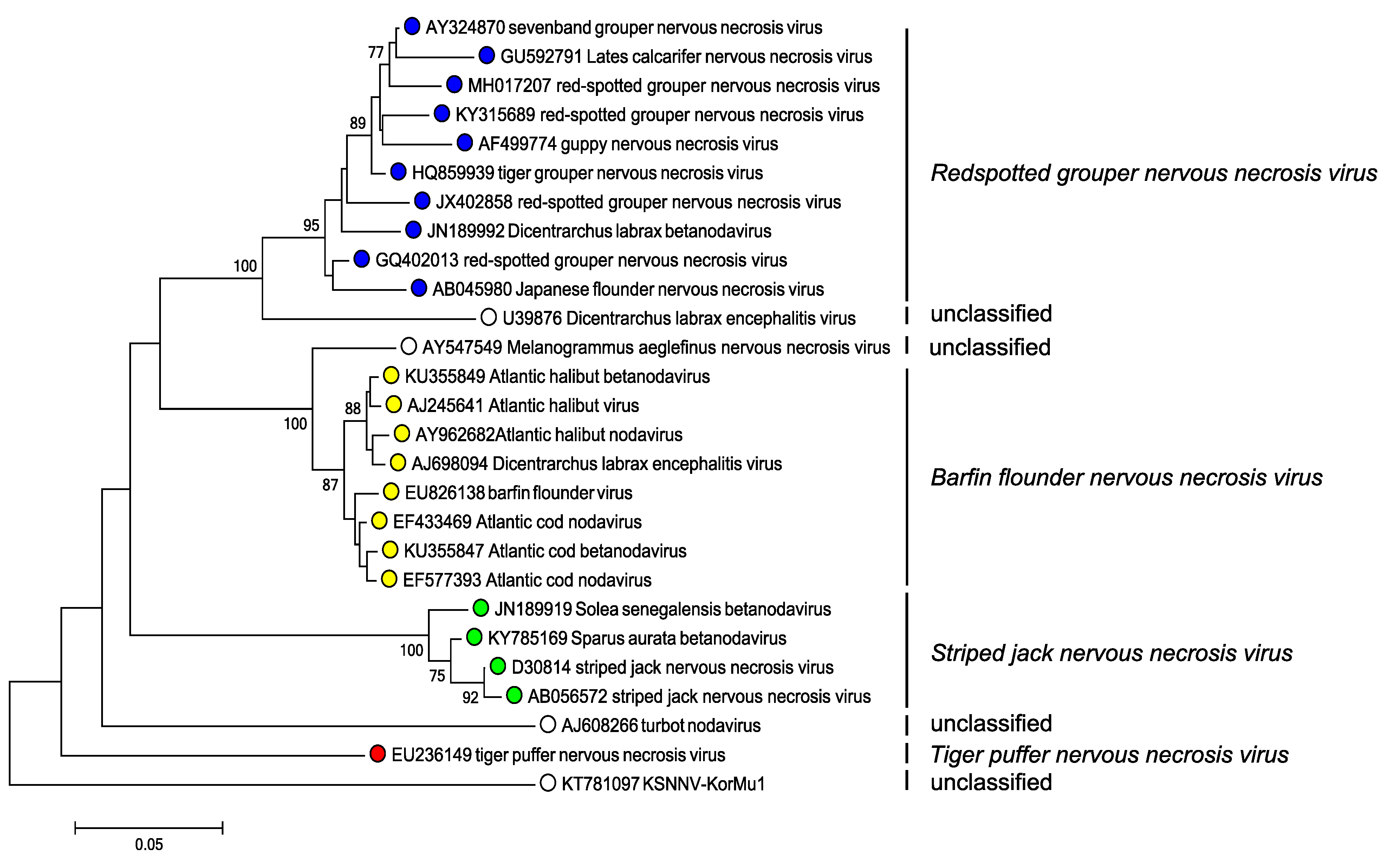 |
| Figure 2. Betanodavirus Phylogenetic analysis of betanodavirus capsid protein sequences. Complete RNA2 sequences were aligned using MUSCLE (Edgar 2004) within the SSE v1.3 (Simmonds 2012). After excluding sequences that were <2% divergent in amino acid sequence, a neighbor joining tree produced using evolutionary distances using the JTT matrix within MEGA7 (Kumar et al., 2016). Branches supported by >70% of bootstrap replicates are indicated. Tips are labelled with nucleotide sequence accession number, virus name and virus species. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
Dicentrarchus labrax encephalitis virus |
RNA2: U39876 |
DlEV |
|
KSNNV-KorMu1 |
RNA2: KT781097 |
|
|
Melanogrammus aeglefinus nervous necrosis virus |
RNA2: AY547549 |
|
|
turbot nodavirus |
RNA2: AJ608266 |
TNV |
Virus names and virus abbreviations are not official ICTV designations.

