Family: Nodaviridae
A. S. Sahul Hameed, Arun S. Ninawe, Toshihiro Nakai, Shau-Chi Chi and Kyle L. Johnson
The citation for this ICTV Report chapter is the summary published as Sahul Hameed et al., (2019):
ICTV Virus Taxonomy Profile: Nodaviridae, Journal of General Virology, 100, 3–4.
Corresponding author: Sahul Hameed, A.S. (cah_sahul@hotmail.com)
Edited by: Nick J. Knowles and Stuart G. Siddell
Posted: October 2018
PDF: ICTV_Nodaviridae.pdf
Summary
The family Nodaviridae includes two genera, Alphanodavirus and Betanodavirus (Table 1. Nodaviridae). The family name derives from the Japanese village of Nodamura where Nodamura virus was first isolated from Culex tritaeniorhynchus mosquitos. Virions are non-enveloped and spherical in shape with icosahedral symmetry (T=3) and diameters ranging from 25–33 nm. The genome consists of two molecules of positive-sense ssRNA: RNA1 and RNA2. The virion capsid consists of 180 protein subunits arranged on a T=3 surface lattice. Alphanodaviruses infect insects whereas betanodaviruses are pathogens of fish.
Table 1. Nodaviridae. Characteristics of members of the family Nodaviridae.
| Characteristic | Description |
| Typical member: | striped jack nervous necrosis virus (RNA1: AB056571; RNA2: AB056572) species Betanodavirus pseudocarangis |
| Virion | Non-enveloped spherical particles, 25–33 nm in diameter, with or without surface projections |
| Genome | Bi-partite positive-sense, ssRNA of 3.1 kb (RNA1) and 1.4 kb (RNA2) with 5ʹ-terminal caps but without poly(A) tails |
| Replication | Cytoplasmic within virus-induced invaginations on the outer mitochondrial membrane |
| Translation | From capped genomic and subgenomic RNAs |
| Host range | Natural hosts are insects (Alphanodavirus) or fish (Betanodavirus) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Magsaviricetes, order Nodamuvirales; two genera, each including four or more species, a total of nine species |
Virion
Virions are non-enveloped, roughly spherical in shape, 25–33 nm in diameter and have icosahedral symmetry (Johnson and Reddy 1998) (Figure 1. Nodaviridae). The capsid of virions consists of 180 protein subunits arranged on a T=3 surface lattice. Each subunit is composed of a single capsid protein (CP) or the two products of its cleavage. Electron microscopy of negatively-stained betanodaviruses shows surface projections; these are not observed in alphanodaviruses.
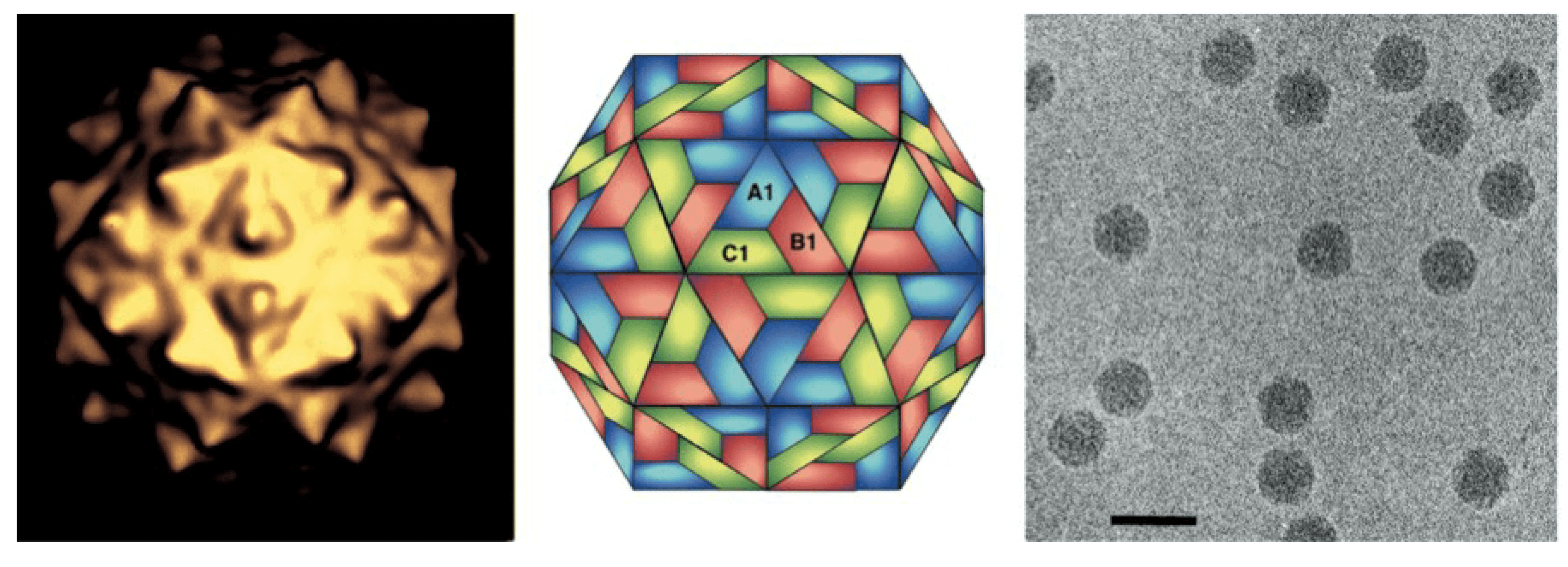 |
| Figure 1. Nodaviridae. Flock House virus particles. (Left) Image reconstruction, (Center) schematic representation of a T=3 icosahedral lattice, A1, B1 and C1 indicate three different quasi-equivalent copies of the capsid protein, (Right) cryo-electron micrograph; the bar represents 50 nm. (Courtesy of N. Olson and T. Baker.) |
Physicochemical and physical properties
Virion Mr is about 8–9 x 106; S20W is about 135–145. Virion buoyant density in CsCl ranges from 1.30 to 1.36 g cm-3. Virions are stable to pH values ranging from 2 to 9 and are resistant to heating at 56 oC for 30 minutes. Infectivity of virions is stable following chloroform extraction.
Nucleic acid
The genome consists of two molecules of positive-sense ssRNA: RNA1 and RNA2. Both RNA molecules are required for infectivity and they are encapsidated in the same virus particle. Both molecules are capped at their 5′-ends and lack poly(A) tails at their 3′-ends. The RNA content of the virion is about 16%. The 3′-ends of alphanodavirus RNAs cannot be chemically derivatized even after treatment with denaturing solvents, indicating that the expected 3′-terminal-OH groups are unreactive.
Proteins
The capsid of virion consists of 180 protein subunits arranged on a T=3 surface lattice. Each subunit is composed of a single CP or the two products of its cleavage.
Lipids
None
Carbohydrates
None
Genome Organization and replication
Virus replication is cytoplasmic. Infected cells contain ssRNAs corresponding to RNA1 and RNA2, as well as the subgenomic RNA3 (387 nucleotides) which derives from the 3′-terminus of RNA1 (Figure 2. Nodaviridae) and is not packaged into virions. In addition to RNA-dependent RNA polymerase (RdRP) activity, protein A binds to and drives invagination of the outer mitochondirial membrane to provide the compartment where RNA replication occurs. RNA3 encodes either one or two proteins; protein B2 (11 kDa) is encoded by all nodaviruses in a reading frame overlapping that of protein A, and is a suppressor of RNA interference. Some nodaviruses also express protein B1 (11 kDa) which corresponds to the C-terminal region of protein A and is of unknown function. Maturation of non-infectious provirions involves the autocatalytic cleavage of protein α into proteins β (39 kDa) and γ (4 kDa).
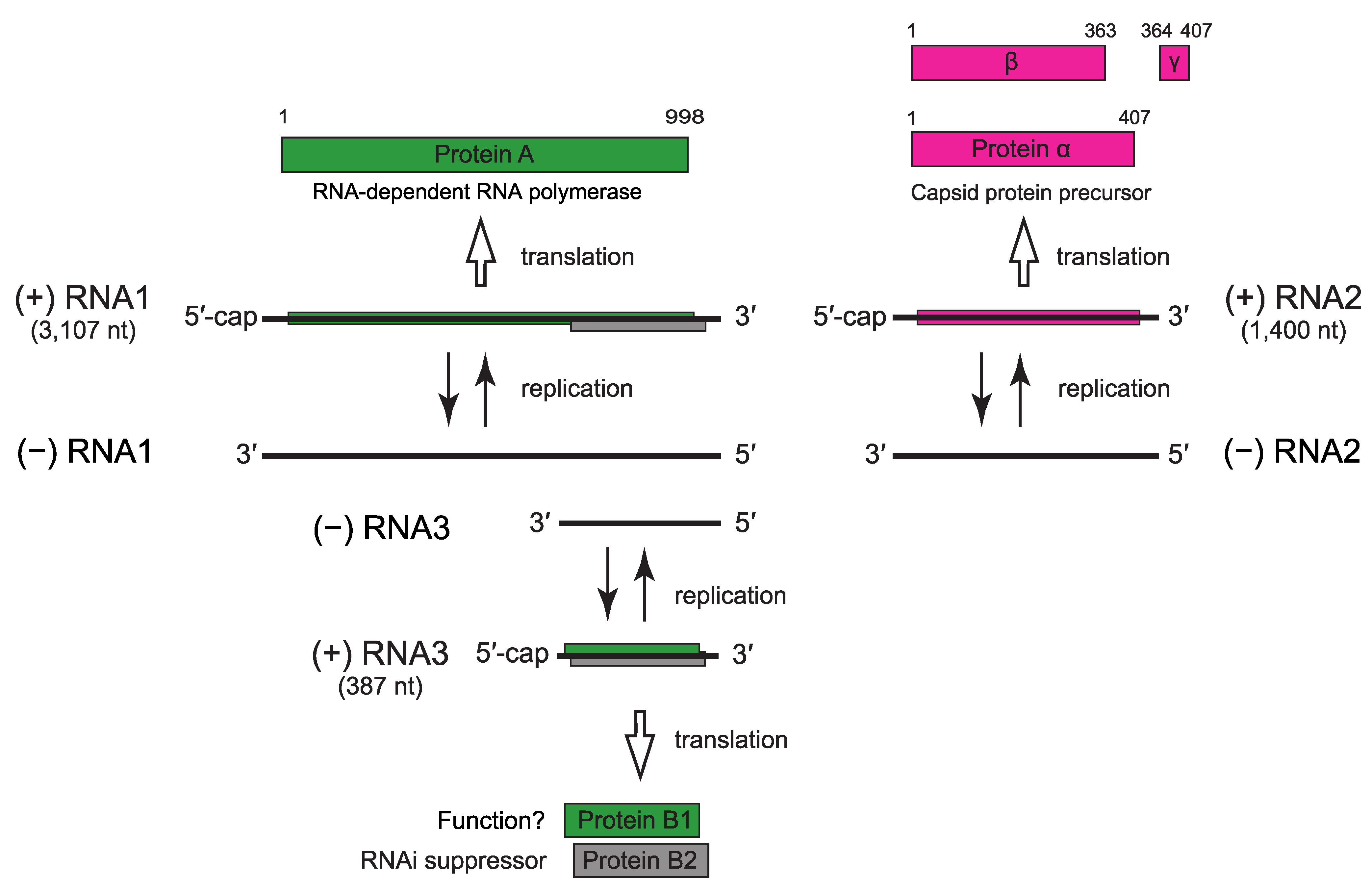 |
| Figure 2. . Nodaviridae. Flock House virus (Alphanodavirus) genome organization and strategy of replication. |
Biology
Members of different species in the family Nodaviridae have been obtained from insects and marine fish (Ball and Johnson 1998, Yong et al., 2017). Some members do not show strict specificity for particular hosts. Virions have both horizontal and vertical modes of transmission that can result in disease in their hosts. Alphanodavirus infection results in stunting, paralysis, and death of its insect hosts, while betanodavirus infection of fish causes neural necrosis, encephalopathy or retinopathy and is associated with behavioural abnormalities and high mortality, posing significant problems for marine aquaculture (Iwamoto et al., 2001, Munday et al., 2002).
Antigenicity
Members of the family Nodaviridae are cross-reactive by immunoblot or double-diffusion immunoprecipitation tests, but all the members represent different serotypes (neutralization titer of each antiserum less than 0.5% in heterotypic crosses). In contrast, some members are not cross-reactive by gel immunodiffusion tests.
Derivation of names
Noda from Nodamura virus the type species of the Alphanodavirus genus, first isolated in Nodamura, Japan.
By convention, species of the Alphanodavirus genus are named after the places of isolation of the exemplar viruses (Boolarra virus, Flock House virus, Nodamura virus and Pariacoto virus), or the host from which the exemplar virus was isolated (Black beetle virus, from the black beetle, Heteronychus arator). Species of the Betanodavirus genus contain viruses that are pathogens of fish causing “viral nervous necrosis” and are, therefore named after common name of the host fish from which they were isolated, followed by nervous necrosis virus, as in Barfin flounder nervous necrosis virus, Redspotted grouper nervous necrosis virus, Striped jack nervous necrosis virus, and Tiger puffer nervous necrosis virus (Iwamoto et al., 2001, Munday et al., 2002).
Genus demarcation criteria
The following criteria can be applied to the demarcation of genera within the family Nodaviridae:
- Biological properties (host range, vectors, mode of transmission)
- Virion physical/physicochemical characteristics (virion sedimentation coefficient and buoyant density)
- Structural protein characteristics (electrophoretic mobilities of the CP precursor or its cleavage products)
- Antigenic properties
- Genome molecular characteristics (in the absence of sequence information, the electrophoretic mobilities of the viral genomic)
- Phylogeny (sequence of the two genomic RNAs, and their predicted proteins).
Relationships within the family
Nodaviruses can be classified based on genetic diversity of the RNA2 segment. Isolates with less than 80% identity at the nucleotide level of RNA2 and less than 87% identity at the amino acid level are classified as different species (Schuster et al., 2014). The CP aa sequences of the alphanodaviruses are only about 10% identical to those of the betanodaviruses.
Relationships with other taxa
The omegatetraviruses such as Nudaurelia capensis omega virus (NCoV) and Helicoverpa armigera stunt virus (HaSV) contain bipartite positive-sense ssRNA genomes, but their RNAs are about twice the size of nodavirus RNAs and they have no 3ʹ-terminal blockage. Tetraviruses also have larger capsids with T=4 icosahedral symmetry (Johnson and Reddy 1998).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Macrobrachium rosenbergii nodavirus | RNA1: AY231436; RNA2: AY222840 | MrNV |
| Penaeus vannamei nodavirus | RNA2: EF137180 | PvNV |
| Wuhan nodavirus | RNA1: AY962576; RNA2: DQ233638 | WhNV |
| Le Blanc nodavirus | RNA2: JQ943580 | LBNV |
| Santeuil nodavirus | RNA2: HM030973 | SNV |
| Orsay virus | RNA2: HM030971 | ONV |
| Mosinovirus | RNA2: KJ632943 | MV |
| Lutzomyia nodavirus | RNA2: KR003800 | LNV |
| bat guano-associated virus | RNA1: HM228873 | BGNV |
| covert mortality nodavirus | KM112247 | CMNV |
| Farfantepenaeus duorarum nodavirus | RNA1: KC441519; RNA2: KC441520 | FdNV |
Virus names and virus abbreviations are not official ICTV designations.
A third group of nodaviruses has been considered by some authors. This group includes the prawn nodaviruses Macrobrachium rosenbergii nodavirus, Penaeus vannamei nodavirus, covert mortality nodavirus and Farfantepenaeus duorarum nodavirus, infecting prawn and shrimp.

