Family: Nodaviridae
Genus: Alphanodavirus
Distinguishing features
Alphanodaviruses have been isolated from insects, although they are capable of infecting cells from a wide range of hosts in the laboratory. They can also infect pigs, mice and hamsters experimentally.
Virion
Virions are non-enveloped, roughly spherical in shape, 32–33 nm in diameter, and have icosahedral symmetry (T=3) (Johnson and Reddy 1998). Electron microscopy revealed no distinct surface structure in the virion. Empty shells are rarely seen in virus preparations (Figure 1. Nodaviridae).
Physicochemical and physical properties
Virion Mr is about 9×106; S20,w is 135–145S. Virion buoyant density in CsCl is 1.30–1.34 g cm−3 and varies with species. The infectivity of aqueous suspensions is stable to extraction with chloroform. The infectivity of Nodamura virus (NoV), black beetle virus (BBV), and Flock House virus (FHV) is stable at room temperature in 1% sodium dodecyl sulfate but Boolarra virus (BoV) is inactivated. Virions are stable at acid pH.
Nucleic acid
The genome consists of two molecules of positive-sense ssRNA: RNA1 (Mr 1.1×106, 3.1 kb) and RNA2 (Mr 0.48×106, 1.4 kb).
Proteins
The capsid consists of 180 protein subunits (protomers) arranged on a T=3 surface lattice. Each protomer is composed of a single capsid protein (CP, protein α) or the two products of its cleavage (proteins β and γ). Mass spectrometry of the FHV CP indicates that the initiating methionine is removed. Thus, for FHV, the capsid proteins are: protein α: (44 kDa), aa 2-407; protein β: (39 kDa), aa 2–363; protein γ: (4 kDa), aa 364–407. Morphogenesis involves the formation of a non-infectious provirion, which acquires infectivity by autocatalytic cleavage of protein α to form proteins β and γ. Maturation cleavage is often incomplete and virions typically contain residual uncleaved protein α.
Lipids
None.
Carbohydrates
None.
Genome organization and replication
Alphanodaviruses replicate in the cytoplasm of infected cells (Figure 2. Nodaviridae). RNA synthesis is resistant to actinomycin D. Infected cells contain three ssRNAs: RNA1 (Mr 1.1×106; 3.1 kb); RNA2 (Mr 0.48×106; 1.4 kb) and a sgRNA3 (Mr 1.10.13×106; 0.39 kb), whose nucleotide sequence corresponds to the 3′-end of RNA1 (387 nt in the case of FHV). Unlike RNAs 1 and 2, RNA3 is not packaged into virions. RNA1 encodes protein A (112 kDa), which is the catalytic subunit of the viral RNA-dependent RNA polymerase (RdRP). RNA2 encodes protein α, the CP precursor (44 kDa). Depending on the virus species, RNA3 encodes one or two small proteins (proteins B1 and B2, 11 kDa). B1 is encoded in the same ORF as protein A. Protein B2 is encoded in an overlapping ORF. The RNA3 of BoV does not encode protein B1 but all known alphanodavirus RNA3 molecules encode protein B2. Protein B2 of FHV functions as a suppressor of RNA interference (RNAi) in Drosophila melanogaster, cultured Drosophila melanogaster cells (Schneider’s line 2), tobacco plants (Nicotiana benthamiana), and the nematode Caenorhabditis elegans. Similarly, NoV B2 suppresses RNAi in cultured mammalian and mosquito cells. The function of protein B1 is unknown. Cells transfected with isolated RNA1 synthesize RNA1 and overproduce RNA3, but do not make RNA2. RNA2 replication strongly inhibits synthesis of RNA3 and the translation of RNA2 suppresses the translation of RNA1 (Johnson et al., 2001).
Biology
Host range
Isolates of all species of alphanodaviruses have been obtained from insects, although serological data suggest that NoV also naturally infects pigs and perhaps herons (Ball and Johnson 1998). NoV seems to be unique among the nodaviruses in its ability to infect and kill both vertebrates and invertebrates. Other alphanodaviruses do not show strict specificity for particular insect hosts.
In the laboratory, most alphanodaviruses can be propagated in larvae of the common wax moth (Galleria mellonella) causing paralysis and death. FHV, BBV, and BoV replicate well in cultured Drosophila melanogaster cells and form plaques on monolayers of these cells. Defective-interfering particles are readily formed unless the viruses are passaged at low multiplicity of infection. FHV, isolated from grass grubs (Costelytra zealandica), also infects several other insect species, including adult common fruit flies (Drosophila melanogaster), tsetse flies (Glossina morsitans morsitans), reduviid bugs (Rhosnius prolixus) and several species of mosquito (Aedes aegypti, Culex pipiens pipiens, Armigeres subalbatus and Anopheles gambiae). FHV can be propagated in mammalian cells, plants (Nicotiana benthamiana), yeast (Saccharomyces cerevisiae) and nematodes (Caenorhabditis elegans). Persistent FHV infections, with subsequent resistance to superinfection, occur readily in cultured Drosophila melanogaster cells. NoV, isolated from mosquitoes, also causes paralysis and death in suckling mice and suckling hamsters. NoV infects cultured cells from both mosquitoes and mammals, but not those of Drosophila melanogaster. Interestingly, NoV infection is delayed in mammalian cells compared to mosquito cells. NoV can also be propagated by transfecting mosquito, vertebrate, or yeast (Saccharomyces cerevisiae) cell cultures with virion RNA or cloned cDNA copies of genomic RNAs at temperatures up to 37 °C. PaV infects cultured cells from the beet armyworm (Spodoptera exigua) corn earworm (Helicoverpa zea), and mosquitoes (Aedes albopictus), but not those of the fruit fly (Drosophila melanogaster) or fall armyworm (Spodoptera frugiperda). Infectious cDNA clones of the genomic RNAs of FHV, NoV and PaV allow the initiation of their respective replicative cycle in many cell types on transfection of plasmid DNA or in vitro transcripts.
Transmission
NoV is transmissible to suckling mice by Aedes aegypti mosquitoes. It causes paralysis and death when injected into suckling mice and suckling hamsters, but no disease in adult animals. In their insect hosts, alphanodaviruses typically cause stunting, paralysis, and death.
Antigenicity
NoV, BBV, FHV and BoV are cross-reactive by double-diffusion immunoprecipitation tests, but all four members represent different serotypes (neutralization titer of each antiserum less than 0.5% in heterotypic crosses). In contrast, Pariacoto virus (PaV) and NoV are not cross-reactive by gel immunodiffusion tests.
Species demarcation criteria
The following criteria can be applied to the demarcation of species within the Alphanodavirus genus:
- Biological properties (host range, vectors, mode of transmission). Since the natural host ranges of the nodaviruses have generally not been examined in detail but may in some cases be broad, virus isolation from a new host is not, in itself, evidence of a new nodavirus species.
- Antigenic properties. Antisera raised against different isolates or strains of a single nodavirus species should exhibit high levels of cross-reactivity in Western blot and/or neutralization analyses. Lower levels of cross-reactivity in these assays using antisera against all previously recognized nodavirus species can provide evidence of a novel nodavirus.
- Virion electrophoretic mobility. Intact virus particles migrate with characteristic electrophoretic mobilities in non-denaturing agarose gel
- Sedimentation coefficient, buoyant density. Virion sedimentation coefficient and buoyant density should be compared with those of other nodavirus species.
- Structural protein characteristics. The electrophoretic mobilities in SDS-PAGE of the CP precursor or its cleavage products should be compared with those of other nodavirus species.
- RNA electrophoretic mobilities. In the absence of sequence information, the electrophoretic mobilities of the viral genomic RNAs should be compared with those of other nodaviruses.
- Phylogeny. Within the alphanodaviruses, CP amino acid sequences are 44–87% identical to one another. Different species encode capsid proteins that differ at > 20% of nucleotide or >13% of amino acid positions. Because the nodavirus genome is segmented, reassortment can occur and the two genome segments may have distinct evolutionary lineages.
In practice, while the five criteria above may be suggestive of a new species, definitive demarcation is based on the nucleotide sequence of the viral CP gene. The two closest recognized species are BBV and FHV, whose RNA2 sequences show 75–78% identity at the nucleotide level and 81–87% identity at the amino acid sequence level (Figure 1. Alphanodavirus). Their RNA1 sequences, however, are 99% identical.
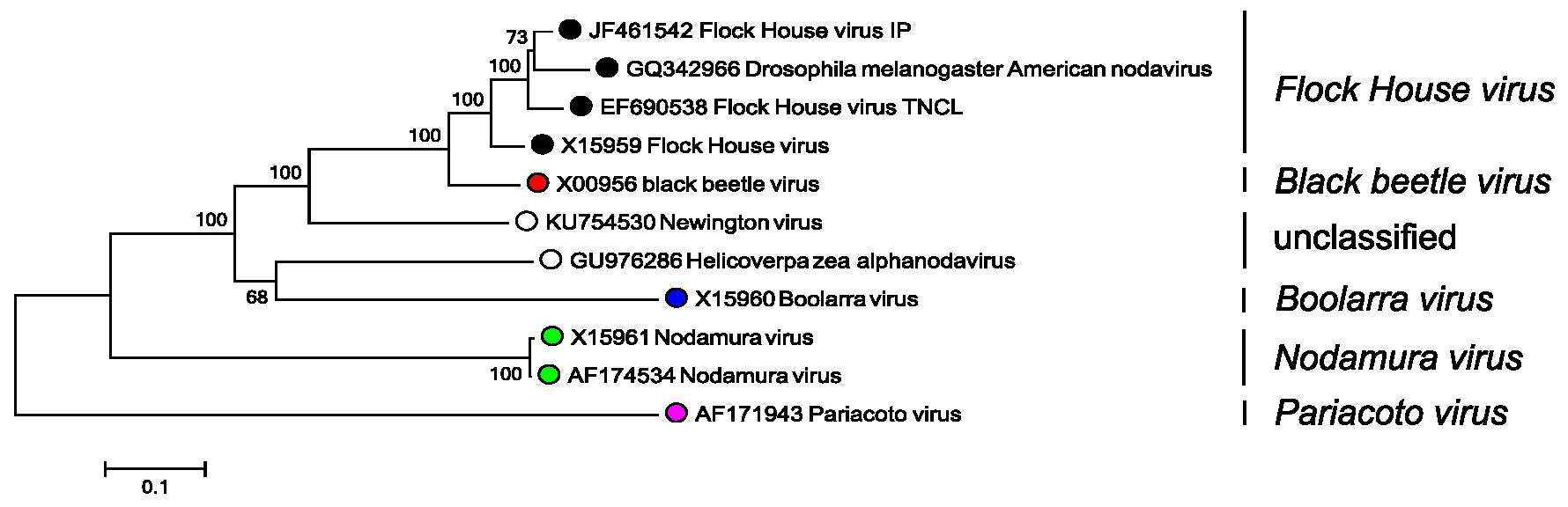 |
| Figure 1. Alphanodavirus. Phylogenetic analysis of alphanodavirus capsid protein sequences. Complete RNA2 sequences were aligned using MUSCLE (Edgar 2004) within the SSE v1.3 (Simmonds 2012) and a neighbor joining tree produced using evolutionary distances using the JTT matrix within MEGA7 (Kumar et al., 2016). Branches supported by >70% of bootstrap replicates are indicated. Tips are labelled with nucleotide sequence Genbank accession number, virus name and virus species. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
Drosophila line 1 virus |
not available |
DL |
|
gypsy moth virus |
not available |
GMV |
|
Helicoverpa_zea_alphanodavirus |
RNA2: GU976286 |
|
|
Lymantria ninayi virus Greenwood |
not available |
LNV |
|
Manawatu virus |
not available |
MwV |
|
New Zealand virus |
not available |
NZV |
|
Newington virus |
RNA2: U754530 |
NV |
Virus names and virus abbreviations are not official ICTV designations.

