Family: Metaviridae
Carlos Llorens, Beatriz Soriano and Mart Krupovic
Corresponding authors: Carlos Llorens (carlos.llorens@biotechvana.com) and Mart Krupovic (mart.krupovic@pasteur.fr)
Edited by: Balázs Harrach and Stuart G. Siddell
Posted: October 2020
PDF: ICTV_Metaviridae.pdf
Summary
Metaviridae is a family of retrotransposons and reverse-transcribing viruses with long terminal repeats (LTRs) popularly referred to as Ty3/Gypsy LTR retroelements in the scientific literature (Table 1. Metaviridae). The Metaviridae belongs to the order Ortervirales together with four other families of evolutionarily related reverse-transcribing viruses. Members of the Metaviridae are widely distributed in eukaryotes and are usually considered to be ancestral to other families of reverse-transcribing viruses, such as Caulimoviridae and Retroviridae. The two genera in the family Metaviridae, Errantivirus and Metavirus, derive from the early discoveries of the Saccharomyces cerevisiae Ty3 virus and its Gypsy-like relatives in drosophilids, which, respectively, differ in the absence or presence of a retroviral env gene encoding the envelope protein. A number of related viruses have yet to be formally classified in order to update and revise classification of metavirids.
Table 1. Metaviridae. Characteristics of members of the family Metaviridae
| Characteristic | Description |
| Typical member | Saccharomyces cerevisiae Ty3 virus (M34549), species Metavirus saccharomycetis |
| Virion | Virions are icosahedral (T = 9) and might be enveloped |
| Genome | Two identical copies of linear single-stranded, positive-sense RNA |
| Replication | Replication by reverse-transcription primed with a host-encoded tRNA |
| Translation | Genomic RNA is translated into one or more polyproteins |
| Host range | Fungi, plants and animals |
| Taxonomy | Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales, family Metaviridae; the two genera Errantivirus and Metavirus include 31 species |
Virion
Morphology
High-resolution structures of immature and mature virus-like particles (VLPs) are available only for Saccharomyces cerevisiae Ty3 virus (SceTy3V) (Dodonova et al., 2019). The Ty3 VLP is an icosahedral particle built on the T=9 lattice (Figure 1. Metaviridae). Structural studies have confirmed the close similarity between the VLPs of metavirids and capsids of retroviruses, consistent with the observation that their capsid and nucleocapsid proteins are homologous (Dodonova et al., 2019, Krupovic and Koonin 2017, Krupovic et al., 2018). As in the case of retrovirids, VLPs of metavirids are essential intermediates in the replication cycle. However, their role in extracellular spread and infectivity of metavirids has been demonstrated only for Drosophila melanogaster Gypsy virus (DmeGypV) (Kim et al., 1994). For other members of the family, the VLPs are primarily or exclusively intracellular (e.g., SceTy3V). Accordingly, the ensemble of VLPs present in the cell is heterogeneous with respect to the stage of maturation, complicating their structural characterization.
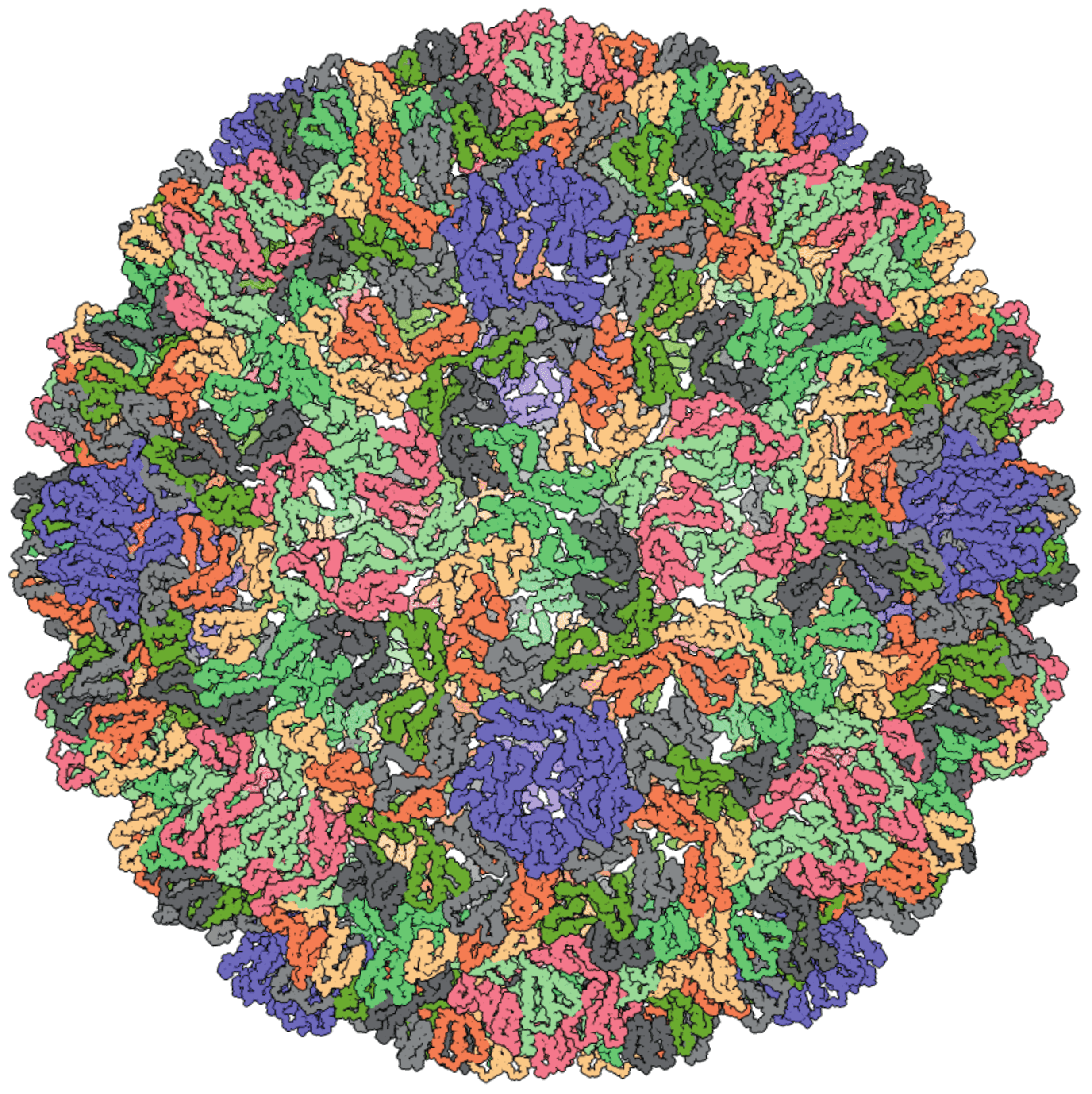 |
| Figure 1. . Metaviridae. Three-dimensional reconstruction of the Saccharomyces cerevisiae Ty3 virus (SceTy3V) virus like particle; both immature and mature particles have the same external radius of about 21 nm, corresponding to a true radius of about 25 nm corresponding to a true diameter of about 47 nm. PDB id: 6R24 (Dodonova et al., 2019). |
Physicochemical and physical properties
In most systems, VLPs are only superficially characterized biochemically.
Nucleic acid
By analogy with retroviruses, it is assumed that Metaviridae VLPs carry two copies of the single-stranded RNA genome. In addition, some cellular RNAs, such as specific tRNAs (usually species specific), involved in the reverse transcription step are also packaged into the VLP.
Proteins
Members of all Metaviridae species normally encode two polyproteins, Gag and Pol. Gag polyprotein is processed into the structural proteins including the capsid protein (CP), which oligomerizes to form the immature VLP, and the nucleocapsid (NC) protein, which is involved in packaging of genomic RNA. The NC of members of most Metaviridae species may have one or more copies of the Cys-X2-Cys-X4-His-X4-Cys (CCHC) zinc finger motif, similar to that observed in the NCs of other viruses in the order Ortervirales (Figure 2. Metaviridae). The exception is provided by errantiviruses which have no apparent NC or CCHC-like feature (Llorens et al., 2009). Finally, most errantiviruses and some metaviruses, such as Arabidopsis thaliana Athila virus (AthAthV) and the Drosophila buzzatii Osvaldo virus (DbuOsvV), encode an envelope polyprotein displaying features of typical transmembrane (TM) and surface (SU) proteins (Wright and Voytas 2002, Pantazidis et al., 1999). However, only the Env polyprotein of errantiviruses has been demonstrated to generate infectious virions (Kim et al., 1994, Pelisson et al., 2002).
Lipids
In the case of members that generate extracellular virions, the viral membrane appears to be derived from the membrane of the host cell.
Carbohydrates
Carbohydrates have not been characterized, although their presence is inferred from sensitivity of the DmeGypV envelope precursor protein to digestion with endoglycosidase F (Song et al., 1994).
Genome organization and replication
The genome of members of the family Metaviridae has two nucleic acid (provirion and RNA genome) states. VLPs contain two copies of positive-sense RNA. The RNA form of the genome is polyadenylated at the 3′-end and serves as a template for transcription, translation and reverse-transcription into cDNA, which is subsequently integrated into the host chromosome as proviral DNA. This proviral DNA is usually flanked by short direct repeats (4–6 bp) of host sequences derived from the insertion site.
The full-length genome of members of the family Metaviridae typically has an internal region flanked by two LTRs which are two homologous non-coding DNA sequences that may begin and end in dinucleotide (5′-TG . . CA-3′) inverted repeats. Genome lengths are variable and may range from 3 kb to more than 15 kb (Figure 2. Metaviridae). LTRs are also variable in size; for example, the LTRs of the Bombyx mori mag virus (BmoMagV) are 77 nt, while those of Drosophila virilis Ulysses virus (DviUlyV) are > 1.2 kb. A canonical LTR has three regions, namely, U3-R-U5, that are analogous to those of retroviruses. U3 is a region of 200–1,200 nt that contains the promoters; “R” is repeated on each end of the transcript; and U5 is a region of 75–250 nt that constitutes the first portion of the reverse-transcribed genome. While LTRs do not contain genes, they carry regulatory elements (enhancers and promoters) that regulate the expression of genes in the internal region of the genome and also, in certain cases, of host genes. The internal region is delimited by two small motifs: a 18 nt sequence downstream to the 5′-LTR (the primer binding site, PBS) and a region of about 10 A/G residues (the polypurine tract, PPT) located upstream of the 3′-LTR.
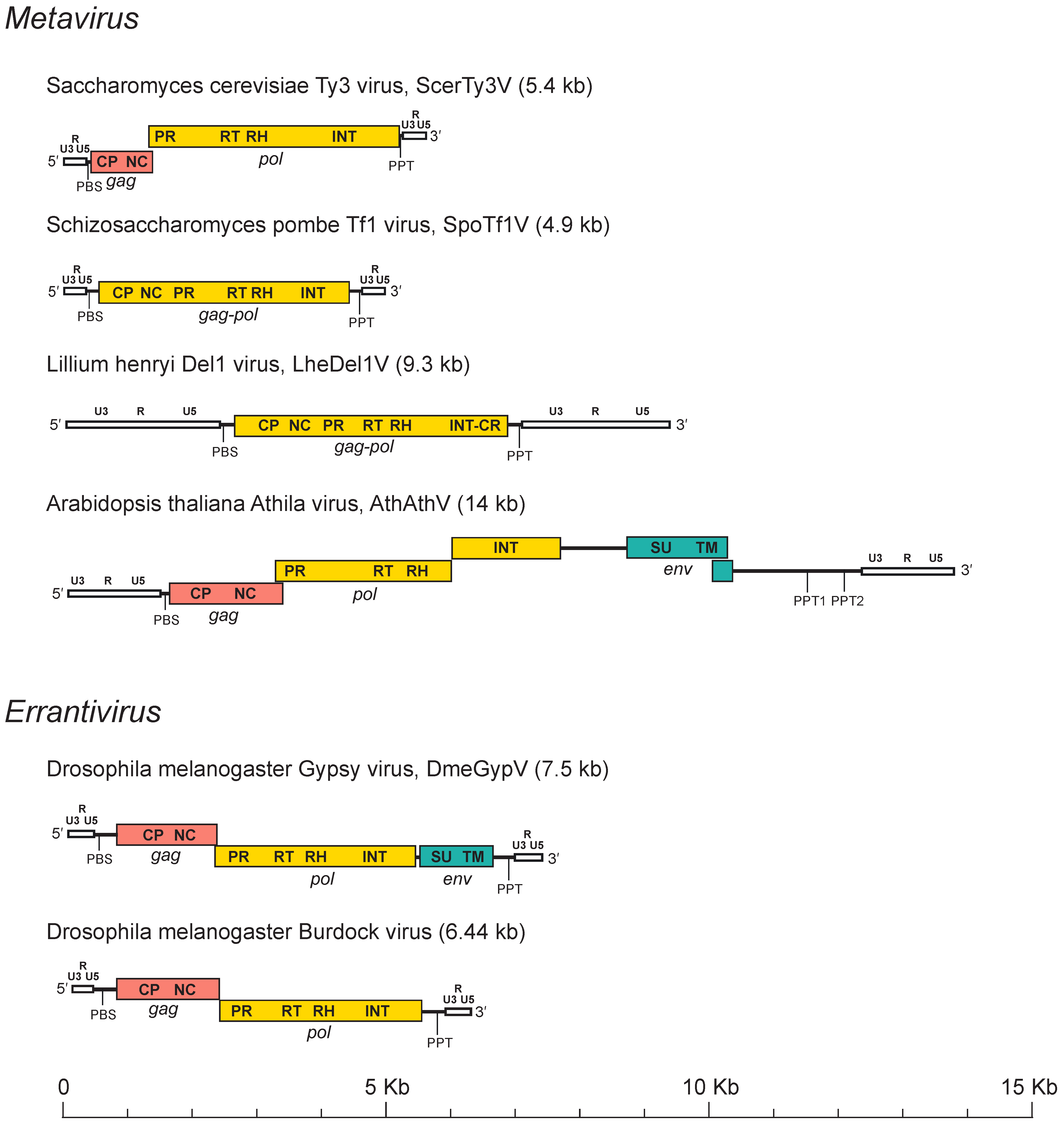 |
| Figure 2. . Metaviridae. Metavirus genome structure. Long terminal repeats (white) are labelled with the U3, R and U5 regions. Other labels are PBS (primer binding site), PPT (polypurine tract), gag (pink) with its capsid (CP) and nucleocapsid (NC) domains, pol (yellow) with its protease (PR), reverse transcriptase (RT), ribonuclease H (RH) and integrase(INT) domains, and env (blue-green) with its surface (SU) and transmembrane (TM) domains. |
The internal coding region may have one gag-pol ORF, two ORFs (gag and pol) or three ORFs (gag, pol and env). Gag encodes domains for the CP and the NC while pol includes domains for the typical protease (PR), reverse transcriptase (RT) ribonuclease H (RH) and Integrase (INT) enzymes required for reverse transcription of the RNA genome into proviral cDNA (within the VLP) and for the integration of the latter into the host genome. PR is the first protein to be cleaved from Pol, although it may also be encoded as a separate protein or as part of Gag protein. PR is a peptidase thought to dimerize, similar to retroviral proteases (Llorens et al., 2008) and that belongs to the clan AA of aspartic peptidases (Rawlings et al., 2018). PR is required for maturation of Gag into CP and NC and for cleavage of Pol into RT-RH and INT proteins. RT is involved in reverse transcription of cDNA from its single-stranded RNA (ssRNA) template and RH is responsible for hydrolysis of the original RNA template that is part of the RNA/DNA hybrid generated during reverse transcription. INT belongs to the retroviral integrase superfamily of DDE-like INTs and transposases (Nowotny 2009) and catalyzes the insertion of reverse-transcribed cDNA into the host genome. Metaviridae INTs present a typical His-X5-His-Xn-Cys-X3-Cys like zinc finger motif followed by the DD35E-like INT core and an additional C-terminal module named GPY/F also present in the INT of viruses in the families Belpaoviridae and Retrovidae (Malik and Eickbush 1999). In addition, INTs of some members of the genus Metavirus infecting plants, fungi and animals contain a conserved chromodomain motif following the GPY/F domain (Malik and Eickbush 1999, Marín and Lloréns 2000). Where characterized, envelope proteins are encoded downstream of INT and are expressed via spliced mRNAs. In almost all members of the Metaviridae, the domain architecture is thus inferred to be: 5′-LTR-CP-NC-PR-RT-RH-INT-LTR3′ for LTR retrotransposons and 5′-LTR-CP-NC-PR-RT-RH-INT-SU-TM-LTR3′ for reverse-transcribing viruses. There are, however, some exceptions to this architecture. For example, a member of the Metaviridae, Gadus morhua Gmr1 virus (GmoGmr1V) has the INT domain upstream of the RT, an arrangement which is characteristic of members of the Pseudoviridae, another family in the order Ortervirales (Butler et al., 2001).
The life cycle of all LTR retroelements consists of four steps: transcription and protein synthesis, RNA packaging and VLP formation, reverse transcription, and integration. Transcription of full-length viral mRNA is initiated by cellular RNA polymerase II from a promoter located in the 5′-LTR. Viral mRNA is then translated into the Gag and Pol polyproteins which are subsequently processed by PR in order to release mature CP, NC, RT, RH and INT proteins required for particle formation, polyprotein maturation, reverse transcription and integration. Note that Pol can be expressed as a translational fusion with Gag or through one or more ribosomal frameshifting events. The mechanism of frameshifting is not uniform among member viruses. In the case of Schizosaccaromyces pombe Tf1 virus (SpoTf1V), the most extensively characterized virus with a single ORF, it appears that a polyprotein is produced and that subsequent proteolytic events are responsible for a high ratio of major structural proteins to catalytic proteins. Little is known about where particle assembly takes place in the cell. Shortly after production of protein precursors, processed polypeptides are observed. By analogy with the closely related retroviruses, it is likely that processing follows, and is dependent upon, intracellular assembly. Reverse transcription takes place in the VLP. In cases where the reverse transcription intermediates have been characterized (DmeGypV, SceTy3V, and SpoTf1V), the cellular tRNA anneals to the RNA genome at the PBS region that is complementary to the 3′-end of that tRNA. The tRNA is used by RT as a primer to synthesize the (-) DNA chain complementary to the R-U5 zone of the 5′-LTR. The tRNA primes the reverse transcription of a short minus-strand species of DNA referred to as the minus-strand-strong-stop (−SSS). As this cDNA is being synthesized, RH degrades the RNA template. Degradation of the 5′-end of the RNA releases the −SSS from the 5′-end of the RNA and permits it to anneal to the R sequence at the 3′-end of the same RNA or of the other co-packaged genome. This permits the −SSS to be extended by RT to generate the minus-strand of the genome. The remainder of the RNA is degraded by RH with the exception of the PPT adjacent to the 3′-LTR, sequence that confers resistance of the RNA fragment to RH cleavage. The PPT serves as the plus-strand primer for reverse transcription. Plus strand cDNA is then synthesized up to the 5′-end of the minus strand, and the PBS sequence of tRNA is used as a template before producing a plus strand strong-stop DNA (+SSS). The +SSS is transferred to the 3′-end of the minus strand previously generated and then, the tRNA primer is removed by RH activity. The complementary sequences of PBS at the 3′-ends of the +SSS and the minus strand anneal with each other to form a circular structure. Each strand serves as a template for RT extension until the full-length double-strand DNA is synthetized and the LTR ends are duplicated. Most recombination events take place during the minus strand synthesis, when the –SSS fragment is transferred from 5′-end of the RNA to R sequence at the 3′-end of a genetically distinct RNA. During retrotransposition the double-stranded proviral cDNA that has been synthetized in the VLP is imported into the nucleus and then inserted into a chromosomal target site, usually generating a 5 bp duplication of host target DNA. Integration is catalysed by the INT protein.
Biology
Depending on the insertion site, integration can be mutagenic if it disrupts or alters gene functions, with potential detrimental effects on the viability of host cells and, by extension, viability of the inserted LTR retroelement. In the course of evolution, members of the Metaviridae (and other retroelements) have developed mechanisms to specifically target integration into loci where alteration of gene function is less likely. This is primarily achieved by integration into noncoding regions through preferential targeting of the heterochromatin regions (that are not permissive for transcription) or by association with centromeric regions. For example, SceTy3V integrates near sites of RNA polymerase III (pol III) transcription by recognizing pol III transcription complexes or chromatin states associated with pol III transcription. By contrast, SpoTf1V recognises certain RNA polymerase II (pol II) promoters. The great majority of errantiviruses are located in the pericentromeric regions of the Drosophila melanogaster genome and this location is conserved in strains from different geographic origins suggesting that a high number of erranviruses were already present in the pericentromeric sequences of the common ancestor of these D. melanogaster strains. In plants, the accumulation of metaviruses, such as AthAthV, Arabidopsis thaliana Tat4 virus (AthTatV) and others, is also observed in centromeric sequences. Previous studies have shown that chromodomain INT proteins encoded by other metaviruses, such as Fusarium oxysporum Skippy virus (FoxSkiV), are able to specify their target site preference by the recognition of characteristic chromatin modifications (Gao et al., 2008). In parallel, host organisms have also developed mechanism to modulate the activity of integrated metaviruses (which may exist in variable DNA methylation states) using distinct types of regulatory elements, such as enhancers, silencers and chromatin insulators. A well-studied example of a chromatin insulator in D. melanogaster is the Gypsy insulator (Wei and Brennan 2001). In contrast, germline activation may also occur for some of these LTR retroelements. For example, SceTy3V transcription is induced by mating pheromone and transposition occurs after mating. In the case of the DmeGypV and the Drosophila melanogaster Zam virus (DmeZamV), transposition occurs in germline cells. All these mechanisms for activation or silencing of the viral genomes are used to contribute to viral latency and escape from the immune system.
Derivation of names
Errantivirus: from Latin errans, “to wander”.
Metavirus: from Greek metathesis for “transposition”, also to connote some uncertainty as to whether these are true viruses or not.
Genus demarcation criteria
The ICTV recognizes two genera within the Metaviridae family - Errantivirus and Metavirus. This classification derives from the original discovery of fungal Ty3-like LTR retrotransposons and their Gypsy-like Env-encoding relatives in drosophilids. According to this original evidence, the distinction between members of the Metavirus and Errantivirus genera is based on the presence or absence of the env gene. Thus, Ty3/Gypsy elements without env are classified as metaviruses, whereas those with env are considered as errantiviruses.
However, this criterion for genus demarcation is inconsistent with the evolutionary history of the Metaviridae. In particular, Ty3/Gypsy lineages with or without true or potential env genes are polyphyletic with respect to each other, suggesting multiple independent acquisitions and losses of the env gene. On the other hand, while the classification of errantiviruses as a genus is well supported in Ty3/Gypsy phylogenies, the criterion for genus demarcation for this genus cannot be solely based on the presence or absence of an env gene since the unclassified errantiviruses HMS-Beagle and Burdock (see (Llorens et al., 2011) and references therein) have the typical LTR-gag-pol-LTR genomic organization, but lack the env gene (Figure 2. Metaviridae). Indeed, an LTR retrotransposon might become a full-fledged virus by acquiring the env gene from another virus. Conversely, a virus may act as an LTR retrotransposon after losing its env gene or the machinery required for infecting other cells. This statement applies to errantiviruses but also to most endogenous retroviruses (ERVs) of vertebrates. Consistent with our arguments, distinct members of the Metavirus genus described in plants and animals have been reported to carry an env gene and thus might have an extracellular phase, although with no demonstrated infectivity. However, these env-carrying metaviruses do not form a monophyletic cluster with errantiviruses and, in fact, due to the high dissimilarity observed among the envelope proteins of these metaviruses and errantiviruses (or other members of the Retroviridae), the simplest hypothesis is that the different lineages carrying env genes acquired their respective env gene independently (Malik et al., 2000).
Relationships within the family
There is broad consensus in the field that the most robust method for assigning taxonomical levels to LTR retroelements (Metaviridae family included) is phylogenetic inference. RT, RH and INT are most commonly used for inferring LTR retroelement phylogenies due to their strong but consistent phylogenetic signal. Although Gag and PR are fast evolving proteins showing high divergence even among members of the same family, they have been shown to produce topologies similar to those for the RT, RH and INT proteins (Llorens et al., 2009, Llorens et al., 2008, Llorens et al., 2011). In the “Resources” section, we provide a link to the Gypsy Database (GyDB) which provides online access to a collection of phylogenetic trees for all reverse-transcribing viruses inferred based on the distinct Gag and Pol (and Env) protein domains. The phylogenetic tree in Figure 3. Metaviridae depicts evolutionary relationships among a representative set of both classified and unclassified viruses representing the currently known diversity of the Metaviridae family. The tree has been inferred based on the most conserved part (the core) of the RT domain and it also includes members of the Pseudoviridae and Belpaoviridae families that, for simplicity, are collapsed as they are used to root the tree. Metaviruses split into distinct phylogenetic lineages, herein referred to as clades or branches which are diverse in terms of the number of OTUs (Operative Taxonomical Units) and host distribution. Interestingly, the phylogenetic signal of all the clades are clearly consistent with a particular distribution of hosts, indicating that the evolutionary dynamics of the Metaviridae viruses is strongly shaped by different adaptive forces involving both intrinsic collaborative/fitness aspects between the lineages constituting each Metaviridae population per genome and the selective pressures provided by the evolution of host and its genomic environment. According to this phylogenetic tree, the genus Errantivirus constitutes a monophyletic clade composed of all Gypsy-like viruses of Drosophila and splits into two clades, Gypsy and Zam, represented by the viruses DmeGypV and DmeZamV, respectively. In contrast to the Errantivirus-Metavirus classification, other members of the family Metaviridae, such as AthAthV and DbuOsvV, carry a third env gene but instead of clustering with errantiviruses, form clades with other lineages. This is, for example, the case of the Athila and Tat clades, represented by viruses AthAthV and AthTatV. Thus, distinct Ty3/Gypsy species formally constituting the genus Metavirus cannot be resolved as a single monophyletic clade and instead show polyphyletic relationships with each other. Of these, the largest and best supported branch is the one comprising Ty3/Gypsy LTR retrotransposons encoding the chromodomain INT proteins (Malik and Eickbush 1999), which are popularly referred to as chromoviruses in the scientific literature (Marín and Lloréns 2000, Gorinsek et al., 2005). The chromoviral branch splits into at least three major clades distributed in plants, fungi and vertebrates, labeled as "Plants" and "Fungi/Vertebrates” in Figure 3. Metaviridae. Another lineage traditionally classified within the genus Metavirus is the Mag clade, a large branch of metazoan Ty3/Gypsy elements, represented by the BmoMagV virus. Other elements assigned to the genus Metavirus are distributed into smaller clades (Cer2-3, Micropia/Mdg3, 412/Mdg1, Osvaldo/GMR1, Kabuki/CsRN1, and others) that in some cases (e.g., Micropia/Mdg1 or 412/Mdg1) are phylogenetically closer to errantiviruses than to other metaviruses.
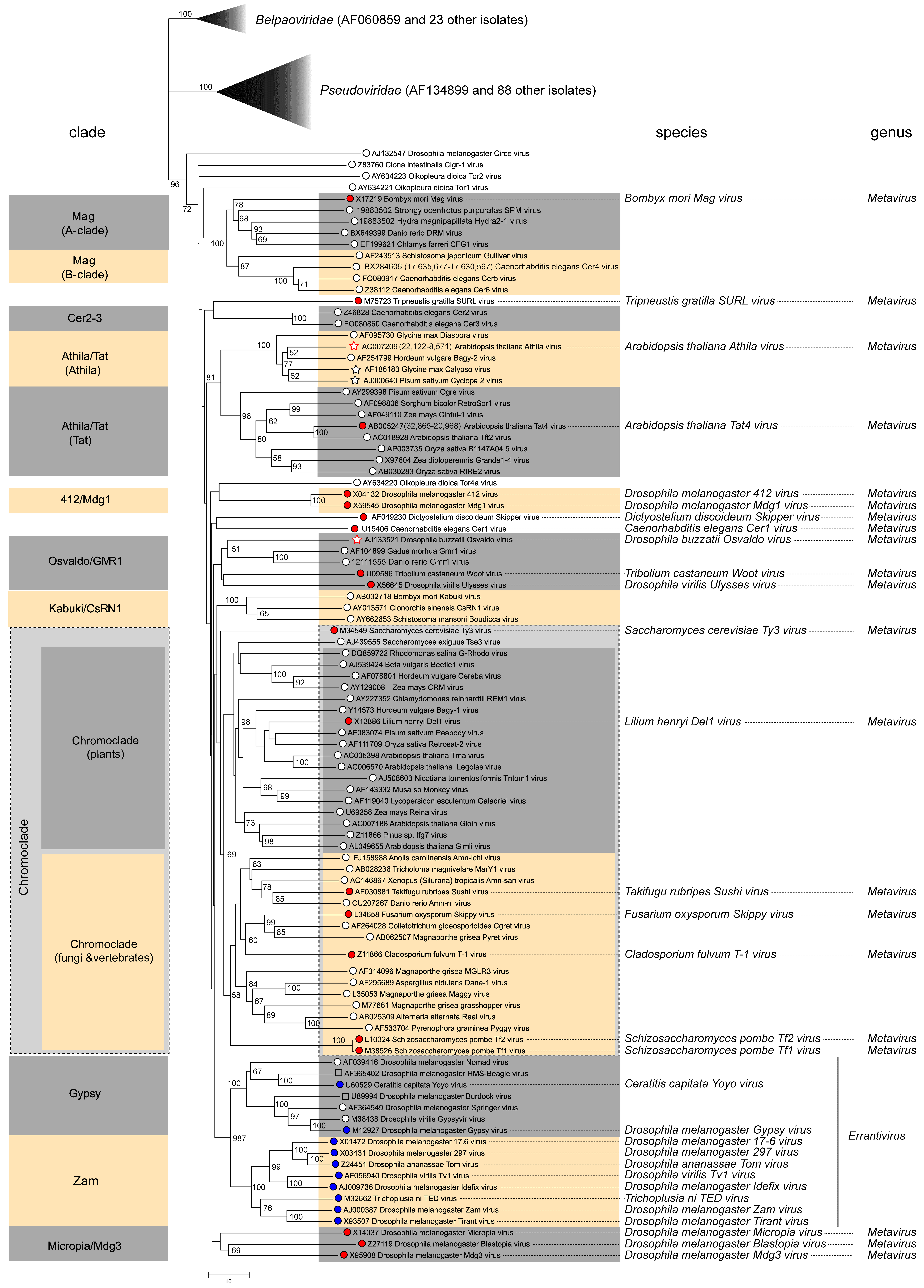 |
| Figure 3. . Metaviridae. Phylogenetic tree of members of the Metaviridae, Pseudoviridae and Belpaoviridae families based on analysis of the RT domain core. Members of the Belpaoviridae and Pseudoviridae families served as outgroups and those nodes are collapsed for clarity. Nodes are labelled with bootstrap values when these were > 70%. Coloured dots and stars indicate isolates belonging to 31 species in the genera Metavirus (red) and Errantivirus (blue), while open circles indicate unclassified viruses in the family. Phylogenetic clades are labelled (left) following the phylogenetic classification of Ty3/Gypsy elements provided at GyDB based on the common phylogenetic signal of both Gag and Pol proteins. Chromoclade refers to the chromodomain-INT-encoding Ty3/Gypsy elements of plants, fungi and vertebrates. Errantiviruses lacking an env gene are indicated with black empty squares, while metaviruses encoding a potential env gene are indicated with empty red or black stars. Tips are labelled with GenBank accession numbers, with coordinates in given in brackets where part of a larger sequence, or where this is not available, with the PMID number of the original publication. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the family Metaviridae share origins and evolutionary history with reverse-transcribing viruses of the families Pseudoviridae, Belpaoviridae, Retroviridae and Caulimoviridae. These five families are unified into the order Ortervirales (Krupovic et al., 2018). In their most basic forms, members of the families Metaviridae, Pseudoviridae and Belpaoviridae are retrotransposons presenting a “LTR-gag-pol-LTR” genomic architecture, albeit certain members of all three families have been shown to carry an env gene, i.e., display the “LTR-gag-pol-env-LTR” genomic architecture typical of simple retroviruses (i.e., those that carry no accessory genes). Thus, the major difference between LTR retrotransposons and simple retroviruses is that the env gene that confers retroviruses their infectious abilities. The relationships of members of the family Caulimoviridae with members of the four other families is even more interesting as it reveals that the conceptual border between a virus and an LTR retrotransposon is extremely flexible and lax. Caulimoviruses are dsDNA viruses infecting plants. Their genome usually presents two ORFs encoding for coat (Gag) and Pol polyproteins, with domain features closely similar to those of LTR retroelements. However, similar to other plant viruses, caulimoviruses encode a movement protein homologous to that found in many plant RNA viruses (Koonin et al., 1991). Consequently, the genomes of caulimoviruses appear to be chimeric, originating through recombination between an LTR retrotransposon, probably a metavirus, and another unknown RNA virus.
Related, unclassified viruses
| Clade | Virus name | Accession number | Coordinates* |
| Athila/Tat (Athila) | Glycine max Calypso virus | AF186183 | |
| Athila/Tat (Athila) | Glycine max Diaspora virus | AF095730 | |
| Athila/Tat (Athila) | Hordeum vulgare Bagy-2 virus | AF254799 | (1–6,244) |
| Athila/Tat (Athila) | Pisum sativum Cyclops-2 virus | AJ000640 | |
| Athila/Tat (Tat) | Arabidopsis thaliana Tft2 virus | AC018928 | (11,471–24,670) |
| Athila/Tat (Tat) | Oryza sativa B1147A04.5 virus | AP003735 | (24,705–46,912) |
| Athila/Tat (Tat) | Oryza sativa RIRE2 virus | AB030283 | |
| Athila/Tat (Tat) | Pisum sativum Ogre virus | AY299398 | |
| Athila/Tat (Tat) | Sorghum bicolor RetroSor1 virus | AF098806 | |
| Athila/Tat (Tat) | Zea diploperennis Grande1-4 virus | X97604 | |
| Athila/Tat (Tat) | Zea mays Cinful-1 virus | AF049110 | |
| Cer2-3 | Caenorhabditis elegans Cer2 virus | Z46828 | (8,185–18,131) |
| Cer2-3 | Caenorhabditis elegans Cer3 virus | FO080860 | (6,211–14,930) |
| Chromoclade | Saccharomyces exiguus Tse3 virus | AJ439555 | |
| Chromoclade (fungi & vertebrates) | Alternaria alternate Real virus | AB025309 | |
| Chromoclade (fungi & vertebrates) | Anolis carolinensis Amn-ichi virus | FJ158988 | |
| Chromoclade (fungi & vertebrates) | Aspergillus nidulans Dane-1 virus | AF295689 | |
| Chromoclade (fungi & vertebrates) | Colletotrichum gloeosporioides Cgret virus | AF264028 | |
| Chromoclade (fungi & vertebrates) | Danio rerio Amn-ni virus | CU207267 | (32,682–38,055) |
| Chromoclade (fungi & vertebrates) | Magnaporthe grisea grasshopper virus | M77661 | |
| Chromoclade (fungi & vertebrates) | Magnaporthe grisea Maggy virus | L35053 | |
| Chromoclade (fungi & vertebrates) | Magnaporthe grisea MGLR3 virus | AF314096 | |
| Chromoclade (fungi & vertebrates) | Magnaporthe grisea Pyret virus | AB062507 | |
| Chromoclade (fungi & vertebrates) | Pyrenophora graminea Pyggy virus | AF533704 | |
| Chromoclade (fungi & vertebrates) | Tricholoma magnivelare MarY1 virus | AB028236 | |
| Chromoclade (fungi & vertebrates) | Xenopus (Silurana) tropicalis Amn-san virus | AC146867 | (28,773–33,738) |
| Chromoclade (plants) | Arabidopsis thaliana Gimli virus | AL049655 | (77,114–82,333) |
| Chromoclade (plants) | Arabidopsis thaliana Gloin virus | AC007188 | (22,521–27,933) |
| Chromoclade (plants) | Arabidopsis thaliana Legolas virus | AC006570 | (38,255–45,793) |
| Chromoclade (plants) | Arabidopsis thaliana Tma virus | AC005398 | (43,771–51,510) |
| Chromoclade (plants) | Beta vulgaris Beetle1 virus | AJ539424 | |
| Chromoclade (plants) | Chlamydomonas reinhardtii REM1 virus | AY227352 | |
| Chromoclade (plants) | Hordeum vulgare Bagy-1 virus | Y14573 | (46,081–59,100) |
| Chromoclade (plants) | Hordeum vulgare Cereba virus | AF078801 | |
| Chromoclade (plants) | Lycopersicon esculentum Galadriel virus | AF119040 | (12,860–19,060) |
| Chromoclade (plants) | Musa sp Monkey virus | AF143332 | |
| Chromoclade (plants) | Nicotiana tomentosiformis Tntom1 virus | AJ508603 | |
| Chromoclade (plants) | Oryza sativa Retrosat-2 virus | AF111709 | (25,889–38,686) |
| Chromoclade (plants) | Pinus sp Ifg7 virus | AJ004945 | |
| Chromoclade (plants) | Pisum sativum Peabody virus | AF083074 | |
| Chromoclade (plants) | Rhodomonas salin G-Rhodo virus | DQ859722 | (13,600–22,168) |
| Chromoclade (plants) | Zea mays CRM virus | AY129008 | |
| Chromoclade (plants) | Zea mays Reina virus | U69258 | |
| Osvaldo/Gmr1 | Danio rerio rGmr1 virus | XM_690885 | (1–3,973) PMID 12111555 |
| Osvaldo/Gmr1 | Gadus morhua Gmr1 virus | AF104899 | (6,448–14,458) |
| Kabuki/CsRN1 | Bombyx mori Kabuki virus | AB032718 | |
| Kabuki/CsRN1 | Clonorchis sinensis CsRN1 virus | AY013571 | |
| Kabuki/CsRN1 | Schistosoma mansoni Boudicca virus | AY662653 | |
| Mag (A clade) | Chlamys farreri CFG1 virus | EF199621 | |
| Mag (A clade) | Danio rerio DRM virus | BX649399 | (60,336–64,818) |
| Mag (A clade) | Hydra magnipapillata Hydra2-1 virus | NW_002123104 | (153,436–158,018) PMID 19883502 |
| Mag (A clade) | Strongylocentrotus purpuratus SPM virus | NW_001353090 | (496–5,740) PMID 19883502 |
| Mag (B clade) | Caenorhabditis elegans Cer4 virus | NC_003284 | (17,635,677–17,630,597) |
| Mag (B clade) | Caenorhabditis elegans Cer5 virus | FO080917 | |
| Mag (B clade) | Caenorhabditis elegans Cer6 virus | Z38112 | |
| Mag (B clade) | Schistosoma japonicum Gulliver virus | AF243513 | |
| Ciona intestinalis Cigr-1 virus | Z83760 | (30,449–34,674) | |
| Drosophila melanogaster Circe virus | AJ132547 | ||
| Oikopleura dioica Tor1 virus | AY634221 | ||
| Oikopleura dioica Tor2 virus | AY634223 | ||
| Oikopleura dioica Tor4a virus | AY634220 |
Virus names and virus abbreviations are not official ICTV designations.
* In column “Coordinates” numbers in parentheses indicate the coordinates of the virus within a larger nucleotide sequence from a host genome. The PMID number of the original report is given when the given Reference sequence has been withdrawn.

