Subfamily: Parahepevirinae
Genus: Piscihepevirus
Distinguishing features
Piscihepeviruses have only been isolated from salmonid fish in North America. Genome organisation is similar to that of orthohepeviruses but piscihepeviruses are phylogenetically distinct (Smith et al., 2014, Batts et al., 2011).
Virion
Morphology
Virions have a diameter of 31 nm (Batts et al., 2011). Other properties of the virion have not been described.
Genome organization and replication
Complete genome sequences are available for 10 strains of cutthroat trout virus as well as one partial genome sequence (haemorrhagic kidney syndrome virus) and 38 partial helicase sequences. The 5′-untranslated region of cutthroat trout virus is longer (100 nt) than has been observed for orthohepevirues (< 35nt). Three open reading frames are present, arranged as for other members of the Hepeviridae (Figure 4.Hepeviridae) except that ORF3 of cutthroat trout virus overlaps the central region of ORF2 rather than locating to its 5′-terminus as is typical of members of the subfamily Orthohepevirinae. The protein encoded by ORF3 has a predicted isoelectric point of 11.8, similar to that encoded by members of the subfamily Orthohepevirinae (12.5), although this reading frame is not conserved in another partial genome sequence that covers this region (AF030878, haemorrhagic kidney syndrome virus). The capsid protein encoded by ORF2 has a predicted isoelectric point of 5.7 with acidic residues clustered at the carboxy-terminus; this contrasts to the basic capsid protein encoded by ORF2 of members of the subfamily Orthohepevirinae (Batts et al., 2011).
Biology
Cutthroat trout virus has been detected in the ovarian fluids of several trout species and is widely distributed in the western United States (Hedrick et al., 1991). A similar virus has been detected in Salmon in New Brunswick, Canada (Kibenge et al., 2000). Although not associated with disease, cutthroat trout virus produces a slow, focal type of cytopathic effect in the Chinook salmon embryo cell line CHSE-214 without destroying the monolayer. Virus can sometimes be re-detected in infected animals after 4-6 weeks. Cutthroat trout virus nucleotide sequences group into two clades (Figure 1.Piscihepevirus), differing by up to 0.27 for either ORF1 or ORF2, considered as representing two genotypes within the single species.
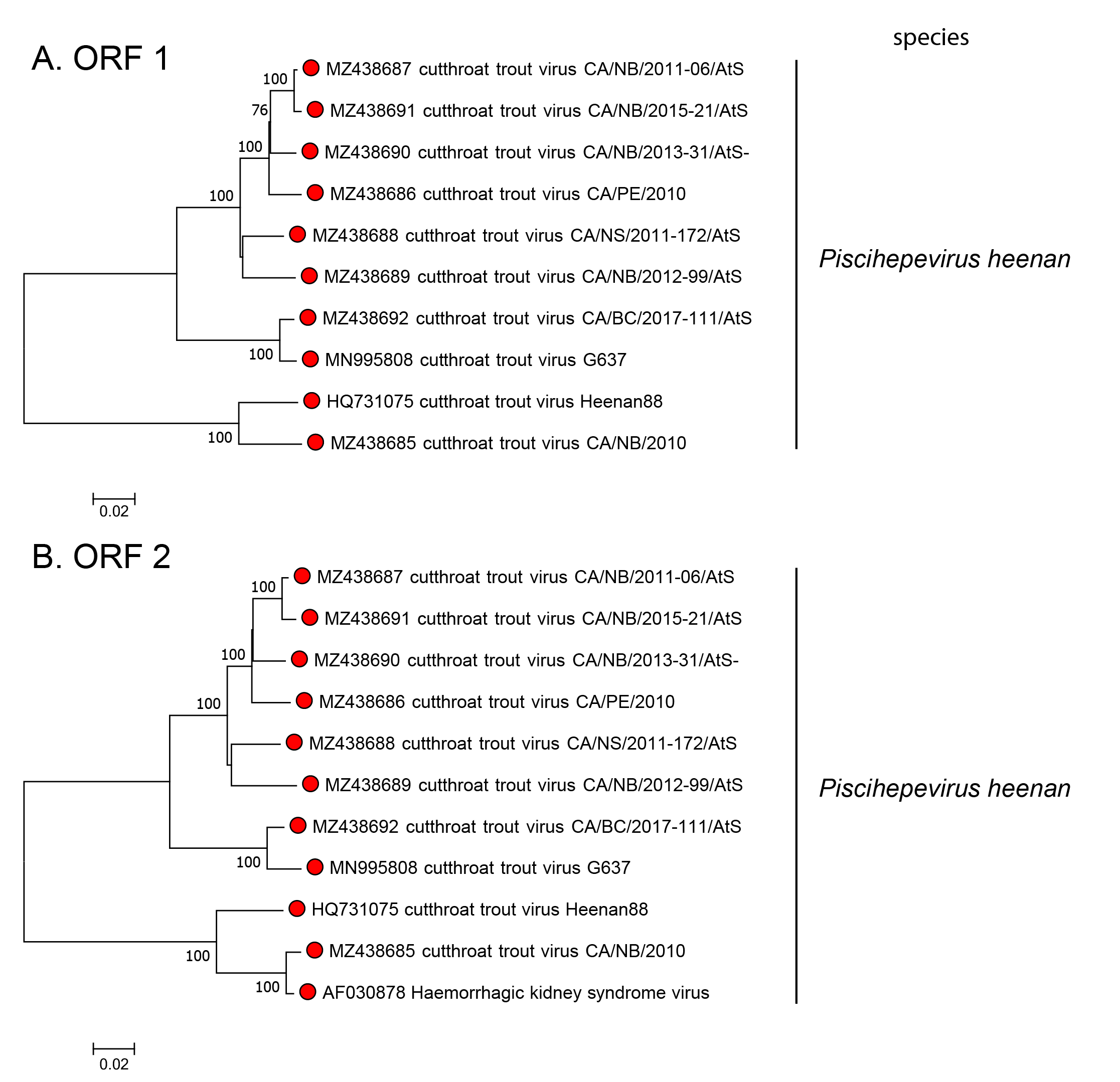 |
| Figure 1.Piscihepevirus Phylogenetic analysis of piscihepevirus sequences. Nucleotide p-distance trees based on alignments of (A) ORF1 and (B) ORF2 sequences using MEGA7 (Kumar et al., 2016). Numbers indicate the bootstrap support for each node (100 replicates). |
Species demarcation criteria
There is only one species in the genus.

