Family: Circoviridae
Genus: Circovirus
Distinguishing features
Circovirus genomes, like cyclovirus genomes, encode at least two major ORFs encoding the replication-associated protein (Rep) and capsid protein (Cp). In the circovirus genomes, the Rep is encoded on the virion sense strand and the Cp on the complementary sense strand, while the gene orientation is opposite in the cycloviruses. Furthermore, the intergenic region at the 3′-end of the ORFs is relatively larger in circovirus genomes than that found in some of the cyclovirus genomes. For the vast majority of cycloviruses, the genes encoding the Rep and Cp overlap at their 3′-ends.
Virion
Morphology
Virions are non-enveloped and have an icosahedral T=1 symmetry (Figure 1. Circovirus). Porcine circovirus (PCV1 and PCV2) and beak and feather disease virus (BFDV) virions range from 15 to 25 nm in diameter (Ritchie et al., 1989, Ritchie et al., 1990, Todd et al., 1991). Structural analysis based on three-dimensional reconstruction of cryo-electron microscopy derived data of PCV2 and BFDV revealed that these circovirus virions have similar appearance with 60 capsid protein subunits arranged in 12 pentameric clusters (Crowther et al., 2003).
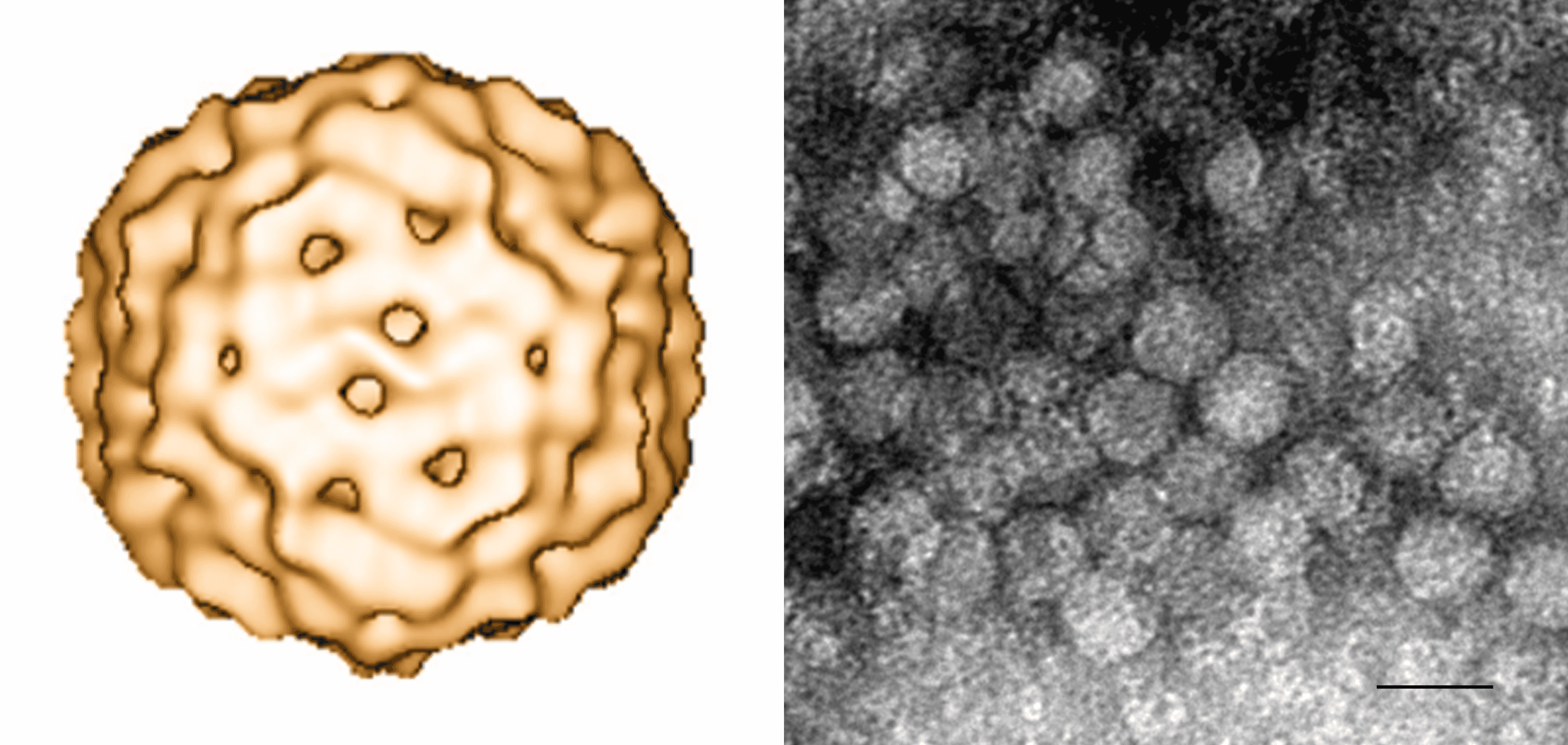 |
| Figure 1. Circovirus. (Left) 3D reconstruction using cryo-electron microscopy of porcine circovirus 2. A structural model comprising 60 subunits (T=1) arranged in 12 pentameric morphological units has been proposed (Crowther et al., 2003). (Right) Negative-stained transmission electron micrograph of porcine circovirus 2 (provided by Carolina Rodríguez-Cariño and Joaquim Segalés, CReSA, Spain). Scale bar = 20 nm. |
Physicochemical and physical properties
The few studies that have investigated physicochemical properties of members of the genus Circovirus suggest that circovirus virions are very stable and resistant to environmental degradation (Todd 2000). PCV1 virions have a sedimentation coefficient of 57S and a buoyant density in CsCl of 1.33–1.37 g cm−3 (Tischer et al., 1982, Allan and Ellis 2000). PCV1 is resistant to inactivation by treatment at pH 3, can withstand incubation at 70°C for 15 min, and is resistant to organic solvents such as chloroform (Allan and Ellis 2000). Haemagglutination assays with BFDV virions concentrated from crude feather suspensions suggest that this avian circovirus has a buoyant density in CsCl of 1.35 g/ml and can withstand chloroform treatment as well as incubation at 80°C for 15 min (Raidal and Cross 1994).
Nucleic acid
Virions contain covalently closed circular ssDNA. The genomes of PCV1 and PCV2 are the smallest viruses shown to replicate autonomously in mammalian cells (1,759 and 1,768 bases). All virus genomes contain a putative ori marked by the conserved nonanucleotide ‘(T/n)A(G/t)TATTAC’ at the apex of a potential stem-loop structure.
Proteins
The virions of PCV1 and PCV2 are each comprised of one structural protein (Cp), for which approximate masses of 36 and 30 kDa have been estimated, respectively (Tischer et al., 1982, Nawagitgul et al., 2000). BFDV virions have been reported to be associated with up to three proteins, 26.3, 23.7 and 15.9 kDa in mass (Ritchie et al., 1990). The protein composition of virions of the other members of the genus Circovirus has not been determined experimentally. However, putative circovirus capsid proteins identified through amino acid similarity searches exhibit conserved patterns of predicted intrinsically disordered regions further supporting their structural function (Rosario et al., 2015).
Lipids
Unknown.
Carbohydrates
Unknown.
Genome organization and replication
Circoviruses possess an ambisense genome organization with the rep gene encoded on the virion strand and the cp gene encoded on the complementary strand of a dsDNA replicative form (RF) (Figure 2. Circovirus). The Rep, which is the most conserved protein amongst members of the genus Circovirus, contains endonuclease and helicase domains involved in RCR. The endonuclease domain, which is located at the Rep N-terminus, contains three conserved sequence motifs: RCR motif I [FT(L/I)NN], RCR motif II [PHLQG] and RCR motif III [YC(S/x)K], with “x” representing any residue (Rosario et al., 2017, Ilyina and Koonin 1992, Rosario et al., 2012). Additionally, the Rep contains a helicase domain towards the C-terminus that is characteristic of superfamily 3 (SF3) helicases encoded by small viruses (Koonin 1993, Gorbalenya and Koonin 1993). The following SF3 helicase motifs are found within a 100–120 amino acid stretch in circovirus Rep primary sequences: Walker-A [G(P/x)(P/x)GxGK(S/t)], Walker-B [uuDDF], and motif C [uTSN], with “x” representing any residue, “u” representing a hydrophobic amino acid, and residues in lower case observed less frequently (Rosario et al., 2017, Rosario et al., 2012). The Cp is much more divergent than the Rep and is only characterized by an N-terminus sequence rich in basic amino acids that may contribute to DNA-binding activity and is expected to interact with packaged viral DNA within virions (Crowther et al., 2003, Niagro et al., 1998).
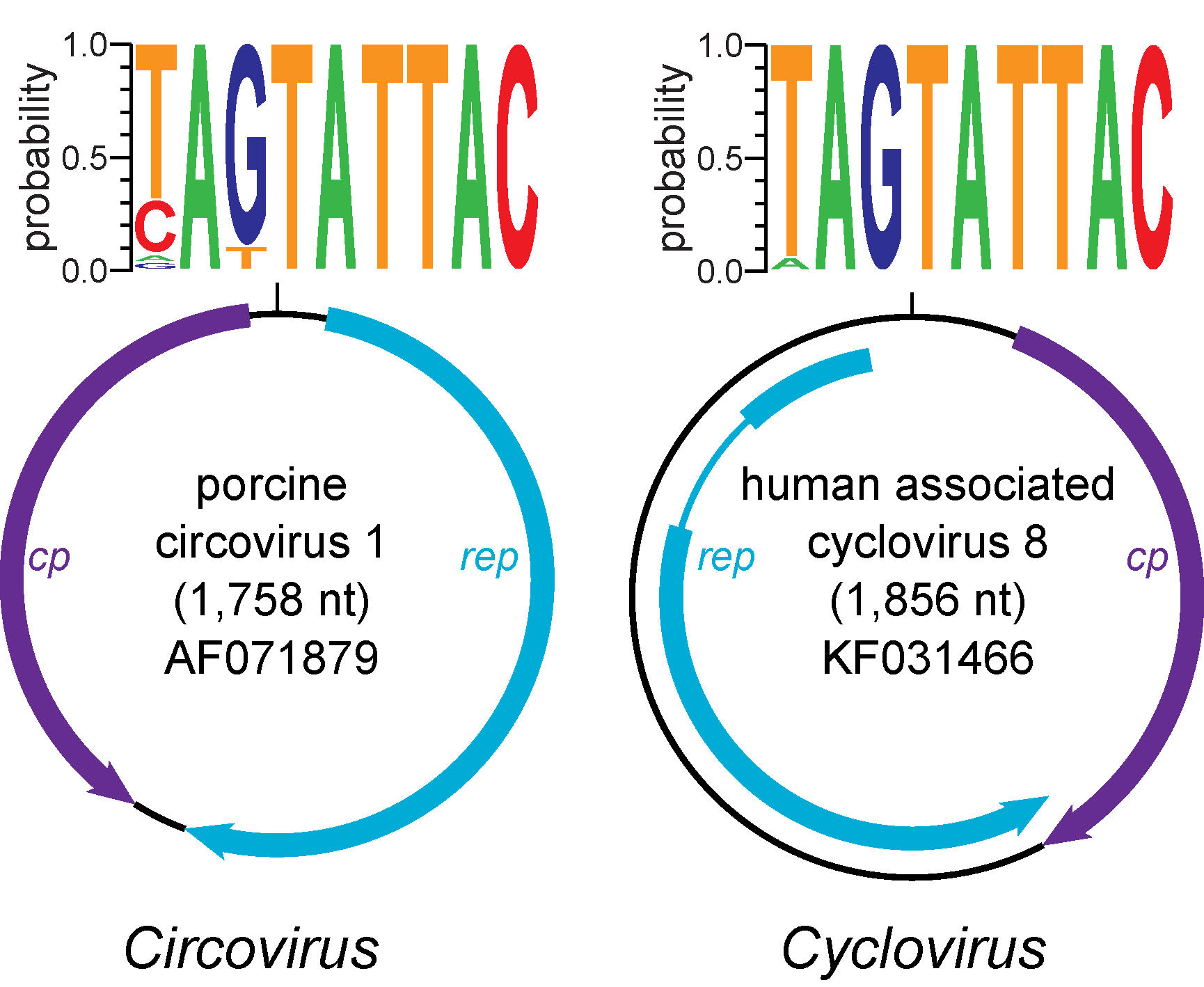 |
| Figure 2. Circovirus. Genome schematics illustrate the major open reading frames (ORFs) characteristic of members of the Circoviridae family. Members of the family Circoviridae, including the Circovirus and Cyclovirus genera, have two major ORFs encoding replication-associated (Rep) and capsid (Cp) proteins as well as a conserved nonanucleotide motif marking the origin of replication. The nonanucleotide motif sequence is depicted through sequence probability logos generated in Weblogo 3 (Crooks et al., 2004). Note that the orientation of major ORFs relative to the nonanucleotide motif differs between genomes representing the Circovirus and Cyclovirus genera. The rep of members of the Cyclovirus type species, Human associated cyclovirus 8 is interrupted by an intron. Although the presence of introns has been observed in various cyclovirus genomes, this has not been reported for circoviruses. Figure reprinted with permission of Springer from Archives of Virology, “Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus”, doi: 10.1007/s00705-017-3247, Rosario, K., Breitbart, M., Harrach, B., Segales, J., Dewart, E., Biagini, P., Varsani, A. © Springer-Verlag Wien 2017 (Rosario et al., 2017). |
Circovirus genomes contain two intergenic regions (IRs), a larger one between the 5′-ends of the two major ORFs and a shorter one between their 3′-ends. The large IR between the initiation codons of the rep and the cp genes contains the ori, which is marked by a conserved nonanucleotide motif ‘NANTATTAC’ at the apex of a stem-loop structure (Mankertz et al., 1997, Cheung 2004a, Rosario et al., 2017, Rosario et al., 2012). The Rep is thought to initiate replication through the RCR model by nicking the virion-sense strand between positions 7 and 8 of the nonanucleotide motif (Steinfeldt et al., 2006). PCV1 replication, presumably through RCR, has been shown to involve production of a dsDNA RF by host DNA polymerases during the S phase of cell division (Tischer et al., 1987).
Transcriptional analyses of PCV1 and PCV2 infecting porcine kidney PK-15 cells have revealed as many as 12 and 9 transcripts, respectively, through alternate splicing (Cheung 2003, Cheung 2004b). Although most of these transcripts are Rep-associated RNAs, only two Rep-associated RNAs, namely Rep and Rep′, have been found to be essential for infectious virus replication (Cheung 2003, Cheung 2004b). The Rep is translated from the full-length transcript (PCV1: 312 aa; PCV2: 314 aa) of the rep gene, whereas a spliced transcript encodes the truncated and C-terminal frame-shifted Rep′ (PCV1: 168 aa; PCV2: 178 aa). Rep and Rep′ of PCV1 bind to two genomic hexameric repeats located close to the potential stem loop spanning the ori (Faurez et al., 2009, Steinfeldt et al., 2001). Both Rep-associated proteins and binding to the hexamers are essential for initiation of replication in these circoviruses (Steinfeldt et al., 2007). In addition to Rep-associated transcripts, porcine circoviruses synthesize a single RNA for the major structural protein Cp (PCV1: 232 aa; PCV2: 233 aa) as well as three RNA transcripts for non-structural proteins with unknown function during infection of PK-15 cells (Cheung 2003, Cheung 2004b). Non-structural proteins, other than Rep and Rep′, have also been shown to play a role in porcine circovirus infections. PCV1 and PCV2 encode a protein known as VP3 that exhibits apoptotic activity (Kiupel et al., 2005, Liu et al., 2005, Hough et al., 2015). In addition, a fourth protein known as ORF4 that exhibits cytoprotection by suppressing caspase activity and inhibiting apoptosis was discovered in PCV2 (He et al., 2013, Lv et al., 2016). Transcriptional information regarding other members of the genus Circovirus remains limited.
Biology
Although members of the genus Circovirus have been identified in various mammals (chimpanzees, dogs, humans, and pigs), birds, and freshwater fish, knowledge of their biology has been largely gathered from porcine circoviruses. Natural infections with PCV1 and PCV2 appear to be restricted to pigs, including various commercial pig breeds and wild boars, while BFDV infections have been detected in over 40 species of psittacine birds (Todd 2000, Allan and Ellis 2000, Baekbo et al., 2012, Raidal et al., 2015). The fecal–oral route of transmission is the most frequent one, although vertical transmission has been reported in cases of PCV2 and avian circovirus infection (Duchatel et al., 2005, Duchatel et al., 2006, Rahaus et al., 2008, Rose et al., 2012, Li et al., 2014). Although there is limited information regarding the transmission of fish circoviruses, detection of barbel circovirus in eggs suggests that these viruses may also be transmitted vertically (Lőrincz et al., 2011). Circovirus infections may cause lymphoid depletion and granulomatous inflammation of lymphoid tissues as well as immunosuppression and are associated with a range of clinical diseases, including infectious psittacine beak and feather disease, circovirus disease of pigeons, and porcine circovirus diseases (PCVDs) in swine, for which PCV2 has been recognized as an essential component of several clinical conditions (Todd 2000, Allan and Ellis 2000, Baekbo et al., 2012, Raidal et al., 2015). However, subclinical circovirus infections are the most common ones.
Antigenicity
PCV1 and PCV2 share common antigenic determinants based on immunoassays against the Rep; however, their Cp is antigenically distinct (Mahe et al., 2000). Porcine circoviruses, including PCV1 and presumably PCV2, are antigenically distinct from beak and feather disease virus (BFDV) (Todd et al., 1991). However, the antigenic relationships among other circoviruses or between avian circoviruses (other than BFDV) and porcine circoviruses have not been explored.
Derivation of names
Circo: from circular conformation.
Species demarcation criteria
The species demarcation threshold is 80% genome-wide nucleotide sequence identity based on the distribution of pairwise identities (Figure 3. Circovirus) (Rosario et al., 2017).
 |
| Figure 3. Circovirus. Distribution of pairwise identities among members of the genus Cyclovirus (purple bars; left) and the genus Circovirus (blue bars; right). Plots reflect pairwise identities based on calculations for the complete genome sequences (top) as well as the replication-associated (rep; middle) and capsid (cp; bottom) genes. All pairwise identities were calculated using the Sequence Demarcation Tool version 1.2 (Muhire et al., 2014) with the MUSCLE alignment algorithm (Edgar 2004). Figure reprinted with permission of Springer from Archives of Virology, “Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus”, doi: 10.1007/s00705-017-3247, Rosario, K., Breitbart, M., Harrach, B., Segales, J., Dewart, E., Biagini, P., Varsani, A. © Springer-Verlag Wien 2017 (Rosario et al., 2017). |

