Family: Circoviridae
Mya Breitbart, Eric Delwart, Karyna Rosario, Joaquim Segalés and Arvind Varsani
The citation for this ICTV Report chapter is the summary published as Breitbart et al., (2017):
ICTV Virus Taxonomy Profile: Circoviridae, Journal of General Virology, 98: 1997–1998.
Corresponding author: Arvind Varsani (arvind.varsani@asu.edu)
Edited by: Balázs Harrach and Peter Simmonds
Posted: August 2017, updated May 2018
PDF: ICTV_Circoviridae.pdf
Summary
The family Circoviridae is comprised of viruses with circular, covalently closed, single-stranded DNA (ssDNA) genomes, including the smallest known viral pathogens of animals (Table 1. Circoviridae). Members of the family are classified in one of two genera, Circovirus and Cyclovirus. Culture- and sequence-independent methods, such as degenerate PCR and metagenomic sequencing, have greatly expanded the known diversity within this family. Viral surveys employing these methods in unconventional hosts have contributed additional members to the genus Circovirus and led to the creation of the genus Cyclovirus, for which there are no cultured representatives. Species assignment to a given genus is based on genome organization, particularly the location of the origin of replication (ori) relative to coding regions. The species demarcation threshold for members of the family Circoviridae is 80% genome-wide nucleotide sequence identity.
Table 1. Circoviridae. Characteristics of members of the family Circoviridae.
| Characteristic | Description |
| Typical member | porcine circovirus 1 (AF071879), species Circovirus porcine1 |
| Virion | Non-enveloped, icosahedral T=1 symmetry, 15–25 nm diameter |
| Genome | Monopartite, circular, single-stranded DNA approximately 1.7–2.1 kb |
| Replication | Rolling circle replication |
| Translation | From at least two mRNAs encoding the replication-associated and capsid proteins |
| Host Range | Members of the genus Circovirus have been identified only in vertebrates (mammals, birds and fish), members of the genus Cyclovirus have been identified in vertebrates (mammals and birds) and invertebrates (arthropods), but in most cases the true host is unknown |
| Taxonomy | Realm Monodnaviria, kingdom Shotokuvirae, phylum Cressdnaviricota, class Arfiviricetes, order Cirlivirales: includes the genera Circovirus and Cyclovirus with 70 and 90 species respectively |
Virion
Morphology
Morphology is only known for some members of the genus Circovirus, which have an icosahedral T=1 symmetry. Virions have not been observed for members of the genus Cyclovirus.
Physicochemical and physical properties
Physicochemical properties have only been investigated for some members of the genus Circovirus. These properties remain unknown for members of the genus Cyclovirus.
Nucleic acid
Virions contain a single molecule of covalently closed, circular ssDNA, which ranges from 1.7 to 2.1 kb.
Proteins
Virion associated proteins have only been identified for some members of the genus Circovirus. Virion proteins have not been investigated for members of the genus Cyclovirus.
Lipids
Unknown.
Carbohydrates
Unknown.
Genome organization and replication
Genomes have ambisense organization containing two major (>600 nt) open reading frames (ORFs), encoding the replication-associated protein (Rep) and capsid protein (Cp) on different strands of a double-stranded DNA (dsDNA) replicative form. Although genome replication is not completely understood for circoviruses and has not been experimentally investigated for cycloviruses, the presence of a conserved Rep and genomic features spanning the putative origin of replication (ori) suggest that members of the family Circoviridae replicate through rolling circle replication (RCR). Studies investigating porcine circovirus replication through recombinant DNA technologies support an RCR model of replication for circoviruses (Mankertz et al., 1997, Cheung 2006, Faurez et al., 2009, Steinfeldt et al., 2007). The Rep is thought to initiate RCR by nicking the virion-sense strand between positions 7 and 8 of a conserved nonanucleotide motif located at the apex of a potential stem-loop structure found in the intergenic region between the 5′-ends of Rep- and Cp-encoding ORFs (Cheung 2004a, Steinfeldt et al., 2006). The nonanucleotide motif marking the ori in genomes from members of the family Circoviridae contains the canonical sequence ‘(T/n)A(G/t)TATTAC’ where lower case nucleotides are observed at low frequency and ‘n’ represents any nucleotide (Rosario et al., 2017).
Biology
All available biological information has been gathered from a few members of the genus Circovirus, mainly porcine circoviruses (PCV1 and PCV2) and beak and feather disease virus (BFDV). There is no biological data regarding the infectivity, transmission, or host range of members of the genus Cyclovirus.
Antigenicity
The antigenic relationships among members of the family Circoviridae are mostly unknown. Porcine circoviruses (PCV1 and PCV2), which share common antigenic determinants, are antigenically distinct from beak and feather disease virus (BFDV). However, the antigenic relationships among other circoviruses have not been explored. Immunoassays have not been performed for members of the genus Cyclovirus.
Derivation of names
Circo: from circular conformation.
Cyclo: from cyclo, latinised form of kyklos in Greek, meaning a circle, wheel or ring
Genus demarcation criteria
Species assignment to a given genus is based on genome organization, particularly the location of the ori relative to coding regions (Rosario et al., 2017). Members of the genus Circovirus have the ori on the same strand encoding the Rep, whereas cycloviruses have the putative ori on the Cp-encoding strand.
Relationships within the family
Members of the genus Circovirus are phylogenetically distinct from members of the genus Cyclovirus (Rosario et al., 2017, Li et al., 2010, Delwart and Li 2012) (Figure 1. Circoviridae). Circovirus genomes detected in mammals are generally more closely-related to each other than to avian circoviruses (Figure 2. Circoviridae). However, circovirus genomes detected in freshwater fish, including barbel and catfish circoviruses, are as distantly related to each other as they are to circoviruses identified in mammals and birds (Lőrincz et al., 2012). Cyclovirus genomes do not cluster by the type of organisms from which they were identified (Figure 2. Circoviridae).
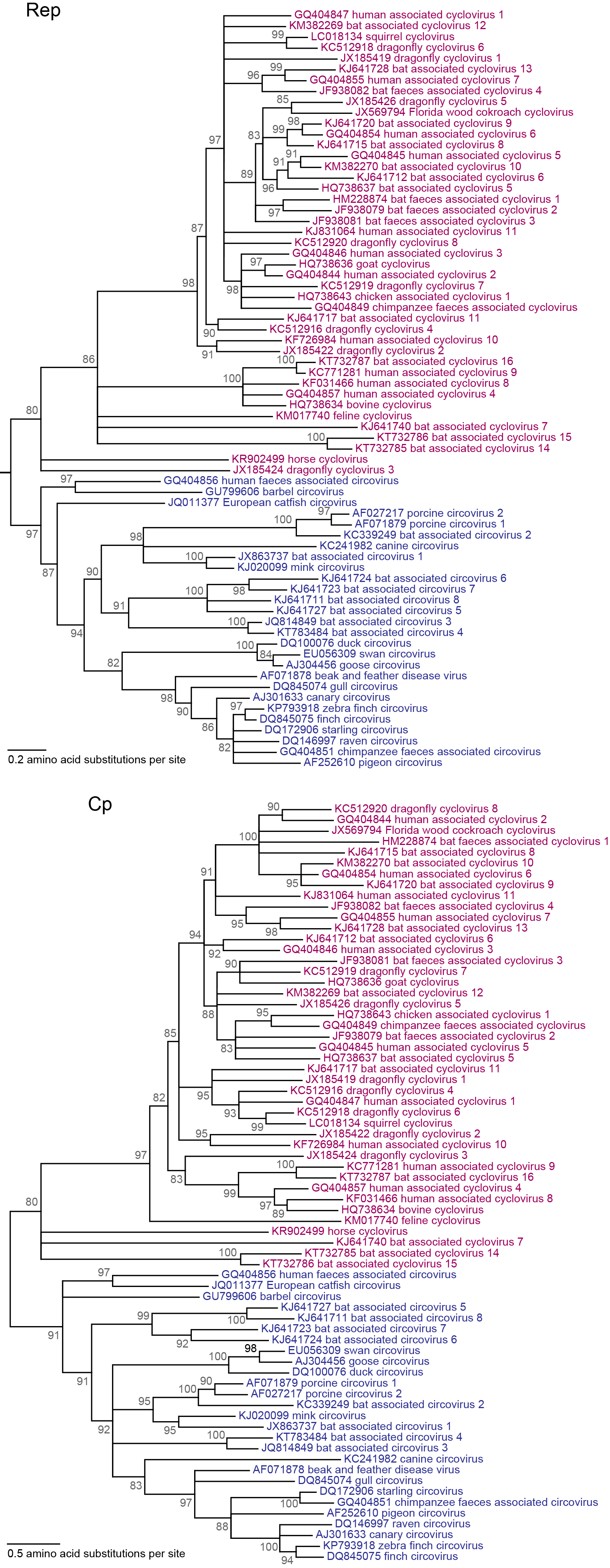 |
| Figure 1. Circoviridae. Maximum likelihood (ML) phylogenetic trees of representative replication-associated protein (Rep; top) rooted with Reps of closely related sequences (GenBank accession numbers JX904473, KRC248418 and KJ547623; rooting sequences not shown) and mid-point rooted capsid protein (Cp; bottom) amino acid sequences of representatives from species within the Circovirus (blue font) and Cyclovirus (purple font) genera. The ML trees were inferred using PhyML (Guindon et al., 2010) with the LG model of substitution after aligning amino acid sequences using the MUSCLE algorithm (Edgar 2004). Branches with <80% SH-like support have been collapsed. Tips are labelled with nucleotide sequence GenBank Accession number and virus name. Figure modified with permission of the authors and reprinted with permission of Springer from Archives of Virology, “Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus”, doi: 10.1007/s00705-017-3247, Rosario, K., Breitbart, M., Harrach, B., Segales, J., Dewart, E., Biagini, P., Varsani, A. © Springer-Verlag Wien 2017 (Rosario et al., 2017). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
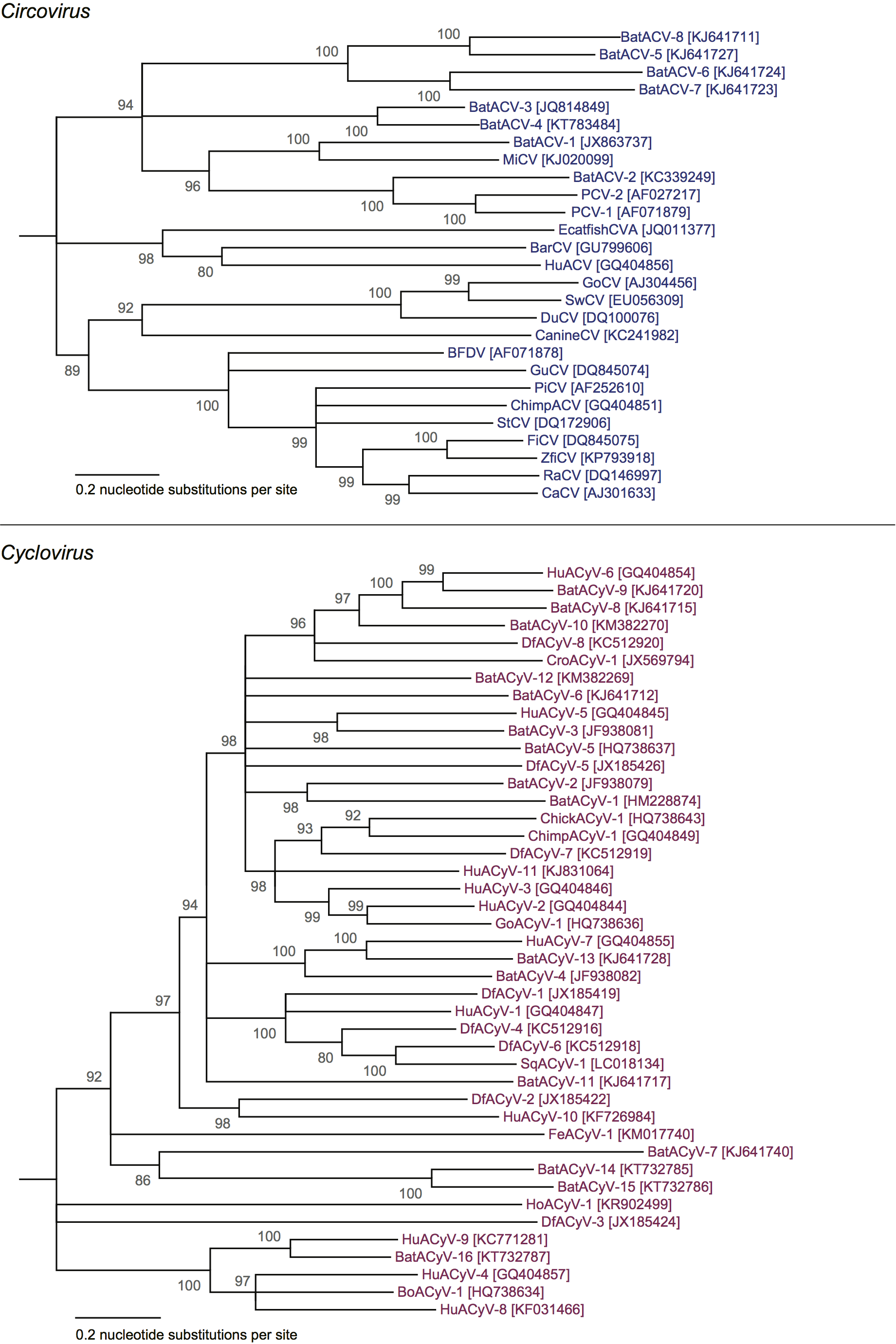 |
| Figure 2. Circoviridae. Maximum likelihood (ML) phylogenetic trees of representative nucleotide sequences from members of species within the Circovirus (top) and Cyclovirus (bottom) genera. The ML trees were inferred using PhyML (Guindon et al., 2010) with the GTR+G model of substitution after aligning complete genome sequences using the MUSCLE algorithm (Edgar 2004). Branches with <80% SH-like support have been collapsed. The phylogenetic trees of circoviruses and cycloviruses were rooted after using cyclovirus or circovirus reverse complemented genome sequences as an outgroup, respectively. Tips are labelled with nucleotide sequence GenBank Accession number and virus name. Figure modified with permission of the authors and reprinted with permission of Springer from Archives of Virology, “Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus”, doi: 10.1007/s00705-017-3247, Rosario, K., Breitbart, M., Harrach, B., Segales, J., Dewart, E., Biagini, P., Varsani, A. © Springer-Verlag Wien 2017 (Rosario et al., 2017). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
The Circoviridae and Anelloviridae families are both comprised of animal viruses with circular single-stranded DNA genomes. The genus Gyrovirus was recently reclassified from the Circoviridae to the Anelloviridae, which includes viruses from a separate lineage of ssDNA replicons based on genomic and structural features (Rosario et al., 2017). Members of the family Circoviridae are evolutionarily related to eukaryotic viruses with circular, ssDNA genomes that encode a Rep with conserved endonuclease and helicase domains (Ilyina and Koonin 1992, Koonin 1993, Rosario et al., 2012, Koonin et al., 2015), including plant-infecting viruses from the families Geminiviridae and Nanoviridae and fungi-infecting viruses from the family Genomoviridae.

