Family: Bornaviridae
Thomas Briese, Ralf Dürrwald, Masayuki Horie (堀江真行), Timothy H. Hyndman, Jens H. Kuhn, Norbert Nowotny, Florian Pfaff, Dennis Rubbenstroth and Keizō Tomonaga (朝長啓造)
The citation for this ICTV Report chapter is the summary published as Rubbenstroth et al., (2021):
ICTV Virus Taxonomy Profile: Bornaviridae, Journal of General Virology 2021 102(7):001613
Corresponding author: Dennis Rubbenstroth (Dennis.Rubbenstroth@fli.de)
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: May 2021, updated December 2022, May 2024
PDF: ICTV_Bornaviridae.pdf (2021 version)
Summary
Members of the family Bornaviridae produce enveloped virions containing a genome consisting of a linear negative-sense non-segmented RNA molecule of approximately 9 kb (Table 1 Bornaviridae). These viruses have been detected in mammals, birds, reptiles, and fish. The family comprises the genera Carbovirus, Cultervirus, and Orthobornavirus. The most-studied viruses with public-health and veterinary impact belong to the species Orthobornavirus bornaense (Borna disease virus 1 [BoDV-1]) and Orthobornavirus sciuri (variegated squirrel bornavirus 1 [VSBV-1]), both of which have proven to cause fatal encephalitis in humans. Several orthobornaviruses cause neurologic and intestinal disorders in birds, mostly parrots. Endogenous bornavirus-like (EBL) sequences have been detected in the genomes of various mammals and other animals of diverse species; these genomic sequences may be remnants of infections by evolutionary relatives of extant bornaviruses.
Table 1 Bornaviridae. Characteristics of members of the family Bornaviridae
| Characteristic | Description |
| Example: | Borna disease virus 1 [U04608], species Orthobornavirus bornaense |
| Virion | Enveloped, spherical virions 90–130 nm in diameter |
| Genome | Linear negative-sense non-segmented RNA of about 9 kb with three transcription units and at least six open reading frames (ORFs) |
| Replication | Intranuclear. Anti-genomic RNA is generated as a replication intermediate that enables synthesis of progeny genomes. Genomic and anti-genomic RNA molecules are neither capped nor polyadenylated |
| Translation | From capped polyadenylated mRNA |
| Host range | Mammals (reservoir: shrews and squirrels; incidental: horses, sheep, humans, and other mammals), birds (parrots, finches, and aquatic birds), reptiles, and fish |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Haploviricotina, class Monjiviricetes, order Mononegavirales: The family includes four genera (Carbovirus, Cartilovirus, Cultervirus, and Orthobornavirus) and 18 species |
The family Bornaviridae includes four genera comprising viruses of 15 species. Bornaviruses are found in a wide range of hosts, including mammals, birds, reptiles, and fish. A unique feature of viruses in the family is the combination of a negative-sense non-segmented RNA genome, the utilization of the cellular splicing machinery in the cell nucleus, and the overlap of transcription units and signals.
Avian Host
Genus Orthobornavirus. Five of the nine species within this genus (Orthobornavirus alphapsittaciforme, Orthobornavirus betapsittaciforme, Orthobornavirus serini, Orthobornavirus estrildidae, and Orthobornavirus avisaquaticae) include viruses that have been detected in birds.
Mammalian Host
Genus Orthobornavirus. Of the nine species within this genus, two comprise viruses that infect mammals: Orthobornavirus bornaense (shrews) and Orthobornavirus sciuri (squirrels). Viruses of both species are of zoonotic importance and can cause fatal nonsuppurative encephalitis in humans.
Piscine Host
Genus Cultervirus. This genus includes one species (Cultervirus hemicultri) for one virus, found in a fish in China.
Reptilian Host
Genus Carbovirus. This genus includes two species (Carbovirus queenslandense and Carbovirus tapeti), each for one virus, found in snakes in Australia.
Genus Orthobornavirus. Two species (Orthobornavirus elapsoideae and Orthobornavirus caenophidiae ) out of the nine in this genus represent viruses found in snakes.
Virion
Morphology
Studies of virions have only been reported for Borna disease virus 1 (BoDV-1), a member of the genus Orthobornavirus. BoDV-1 virions have a spherical morphology and are produced with a bimodal size distribution of two populations of larger (90−130 nm in diameter) and smaller particles(50–90 nm); the population of smaller particles may represent immature or defective particles. Virions are enveloped, appear to carry small spikes (7 nm) and are believed to bud from host-cell membranes (Zimmermann et al., 1994, Kohno et al., 1999).
Physicochemical and physical properties
Properties are only known for BoDV-1 (genus Orthobornavirus). Partially purified BoDV-1 infectious particles have a buoyant density of 1.16–1.22 g cm−3 in cesium chloride, 1.22 g cm−3 in sucrose, and of 1.13 g cm−3 in renografin. Virus infectivity is lost after 30 minutes of heat treatment at 56 °C. Virions are relatively stable for 3 days at 37 °C , but infectivity loss is observed after 3 days at 37 °C. At 20 °C, infectivity is lost after 4 weeks and at 4 °C after 4–6 months. The virus is stable at -70 °C for at least 2 years, whereas there is approximately a 100-fold loss of infectivity when stored at -20 °C. Virions are inactivated below pH 5.0 or by treatment with organic solvents, detergents, or exposure to ultraviolet radiation. Infectivity is destroyed by lipid solvents (Danner and Mayr 1979).
Nucleic acid
Bornaviruses possess linear negative-sense non-segmented RNA genomes of approximately 9 kb.
Proteins
Bornaviruses express five structural proteins—nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), the large protein (L) containing an RNA-directed RNA polymerase domain—and one accessory protein (X). More detailed characteristics are only known for members of the genus Orthobornavirus (see Table 1.Orthobornavirus in the Orthobornavirus genus section).
Lipids
No details on the composition of the bornaviral envelope are reported.
Carbohydrates
Studies have only been reported for BoDV-1 (genus Orthobornavirus). The surface glycoprotein G and its post-translational derivatives carry high-mannose N-glycans and mixtures with complex-type glycans, respectively (Kiermayer et al., 2002).
Genome organization and replication
Bornaviruses have genomes consisting of a negative-sense non-segmented single-stranded RNA approximately 9 kb in length that is organized into three transcription units and at least six open reading frames (ORFs) in the order 3′-N-X/P-G-M-L-5′ for carboviruses and culterviruses or 3′-N-X/P-M-G-L-5′ for orthobornaviruses (Figure 1 Bornaviridae) (Kuhn et al., 2015, Hyndman et al., 2018, Shi et al., 2018).
Modes of virus replication are primarily known for members of the genus Orthobornavirus (see “Genome organization and replication” section in the Orthobornavirus genus section).
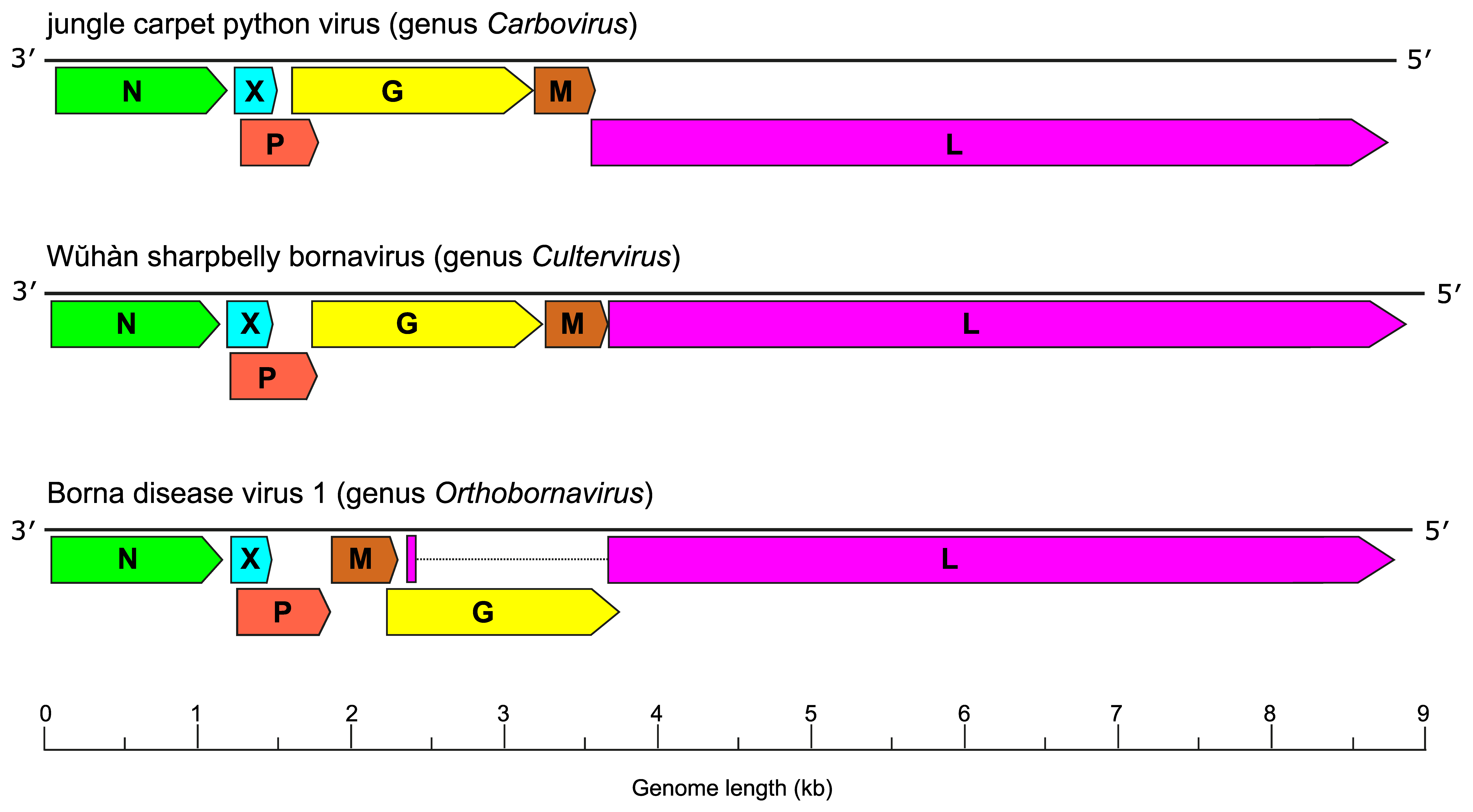 |
| Figure 1 Bornaviridae. Genome organization of viruses in the genera Carbovirus, Cultervirus, and Orthobornavirus. ORFs (depicted on the negative-sense strand) encode N: nucleoprotein; X: accessory protein; P: phosphoprotein; M: matrix protein; G: glycoprotein, and L: large protein containing an RNA-directed RNA polymerase domain. |
Biology
Bornaviruses infect a wide spectrum of hosts, including mammals, birds, reptiles, and fish. Since carboviruses and culterviruses have only been characterized genetically, information is mainly available for the members of genus Orthobornavirus (see “Biology” section in the Orthobornavirus genus section).
Antigenicity
Mainly known for members of the genus Orthobornavirus (see “Antigenicity” section in the Orthobornavirus genus section).
Genus demarcation criteria
Criteria for bornavirus genus demarcation are based on genomic characteristics—including PAirwise Sequence Comparison (PASC) (Bao et al., 2012, Bao et al., 2014), phylogeny, and genome organization. The genus-demarcation cut-off was set at approximately 45% complete genome nucleotide identity (Kuhn et al., 2017, Rubbenstroth et al., 2018).
Derivation of names
Bornaviridae: named after its first member, Borna disease virus 1 (BoDV-1). “Borna disease” can be traced back to a frequent occurrence of fatal encephalitis in horses on farms in the district around the city of Borna in Western Saxony (Germany) in the late 19th century. The term was widely used by farmers at that time and later adopted by scientists.
Carbovirus: from “Cairns”, the origin of the snake in which a carbovirus was first detected, “reptilian”, and “bornavirus”.
Cultervirus: from the binomial name of the virus host Hemiculter leucisculus (Basilewsky, 1855), wild carp.
Orthobornavirus: from the Greek ὀρθός (orthós), which means “correct”; stands for the fact that viruses of this genus were the first viruses of the family Bornaviridae to have been detected (e.g., BoDV-1).
Relationships within the family
Phylogenetic relationships across the family have been estimated using maximum-likelihood trees generated from complete and partial protein sequences (Figure 2 Bornaviridae). Note that all three genera are related to endogenous sequence elements integrated into the genomes of various animals (see “Related, unclassified viruses”).
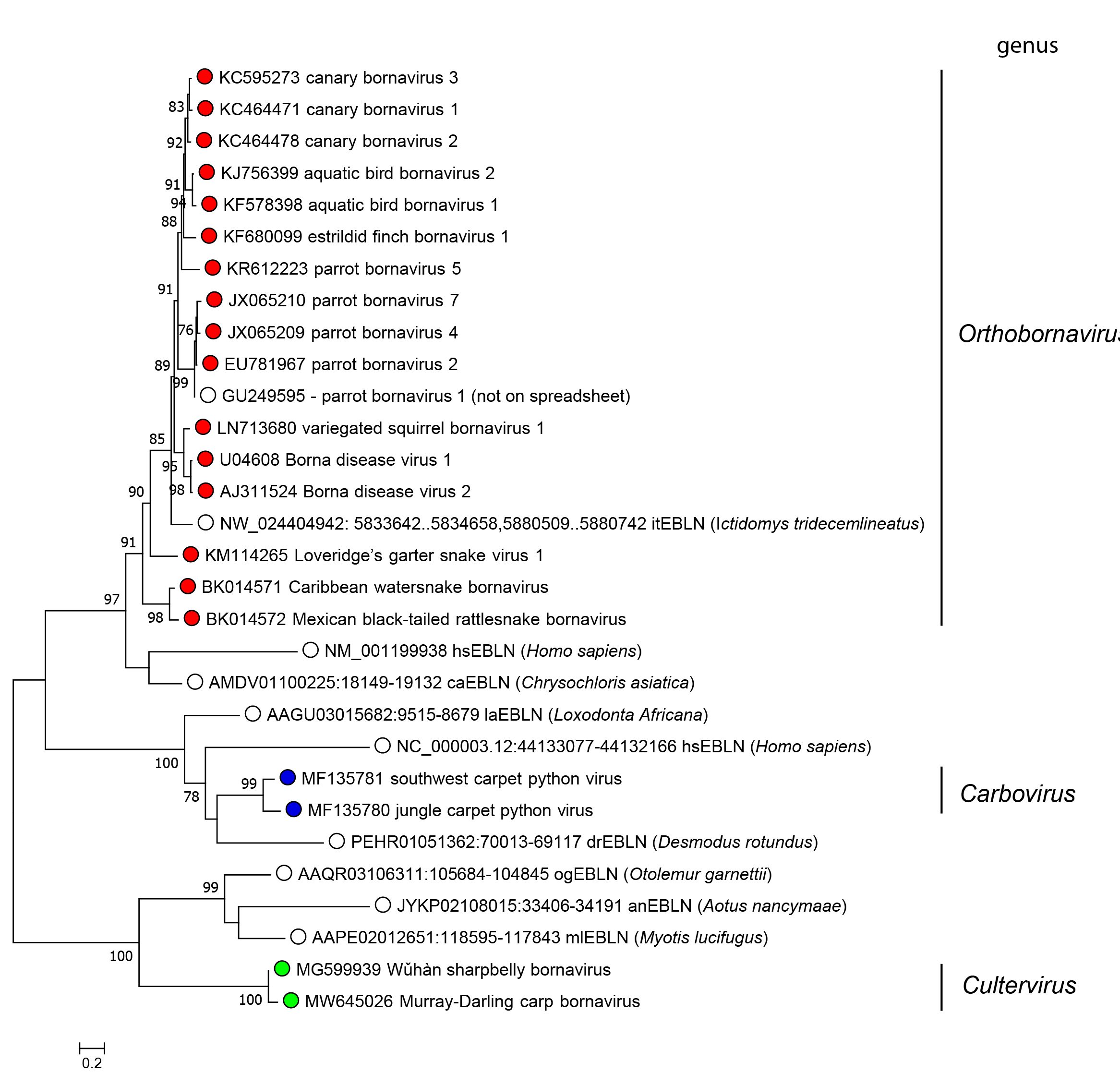 |
| Figure 2 Bornaviridae. Phylogeny based on alignment of bornavirus nucleoprotein and endogenous bornavirus-like N sequences. The tree presents putative N amino-acid sequences of members of different Bornaviridae genera, represented by coloured circles, and endogenous bornavirus-like N sequences, represented by open squares. The sequences were aligned by L-INS-i algorithm in Multiple Alignment using Fast Fourier Transform (MAFFT) version 7.407. The tree was inferred using the maximum-likelihood method and pruned with an option for partial deletion of 90% and an LG+G model, determined by a DNA/protein model search function using Molecular Evolutionary Genetics Analysis Version 10 (MEGA X) software (Kumar et al., 2018). The reliability of the tree was assessed by 100 bootstrap replicates with values over 70% shown. |
Relationships with other taxa
Bornavirus genomic organization is similar to that of other negative-sense non-segmented RNA viruses in the order Mononegavirales. Bornavirus genomes (about 9 kb) are considerably shorter than those of the other known members of the order Mononegavirales: Artoviridae (about 12 kb), Filoviridae (about 19 kb), Lispiviridae (10–14 kb), Mymonaviridae (about 10 kb), Nyamiviridae (10–12 kb), Paramyxoviridae (about 15 kb), Pneumoviridae (13–15 kb), Rhabdoviridae (11–15 kb), Sunviridae (about 17 kb), and Xinmoviridae (11–14 kb).
Bornavirus genome expression is likely to be regulated by a combination of overlapping transcription units, overlapping of ORFs, readthrough of transcription termination signals, and differential use of translational initiation codons. In addition, bornaviruses exploit the nuclear splicing machinery to express their genes, a rare feature among currently known negative-sense non-segmented RNA animal viruses. There is a precedent for the use of each of these strategies by other members of the order Mononegavirales. However, bornaviruses’ concurrent use of such a diversity of strategies for the regulation of gene expression is unique. Phylogenetically, bornaviruses appear most closely related to members of the families Nyamiviridae and Xinmoviridae.
Related, unclassified viruses
Additional unclassified bornaviruses that are probable members of existing genera are listed under individual genus descriptions, as appropriate.
Endogenous bornavirus-like (EBL) elements have been identified in the genomes of various chordates including mammals, birds, reptiles, and fish. These endogenous N, P, M, G, and L sequences are usually truncated or non-translatable and may be remnants of ancient bornaviruses (Horie et al., 2010, Horie et al., 2013). Some EBL elements can be dated to almost 100 million years ago based on their host phylogeny (Kawasaki et al., 2020). Most are distantly related to one of the three known bornavirus genera, but only a few sequences cluster within the diversity of currently circulating bornaviruses (Figure 2.Bornaviridae) (Hyndman et al., 2018).

