Family: Bornaviridae
Genus: Orthobornavirus
Distinguishing features
Viruses in the genus Orthobornavirus are known to infect mammals and birds of numerous species, and one orthobornavirus has been linked to an African reptile (Stenglein et al., 2014, Kuhn et al., 2015). The genomes of orthobornaviruses differ from those of carboviruses and culterviruses in their M and G gene arrangement; in orthobornaviruses M precedes G whereas in members of the other genera it follows G. Because carboviruses and culterviruses have not been isolated in culture and no serologic studies have been published, distinguishing antigenic features are not known.
Virion
Morphology
Borna disease virus 1 (BoDV-1) virions have a spherical morphology (Figure 1.Orthobornavirus). See information provided in the family section.
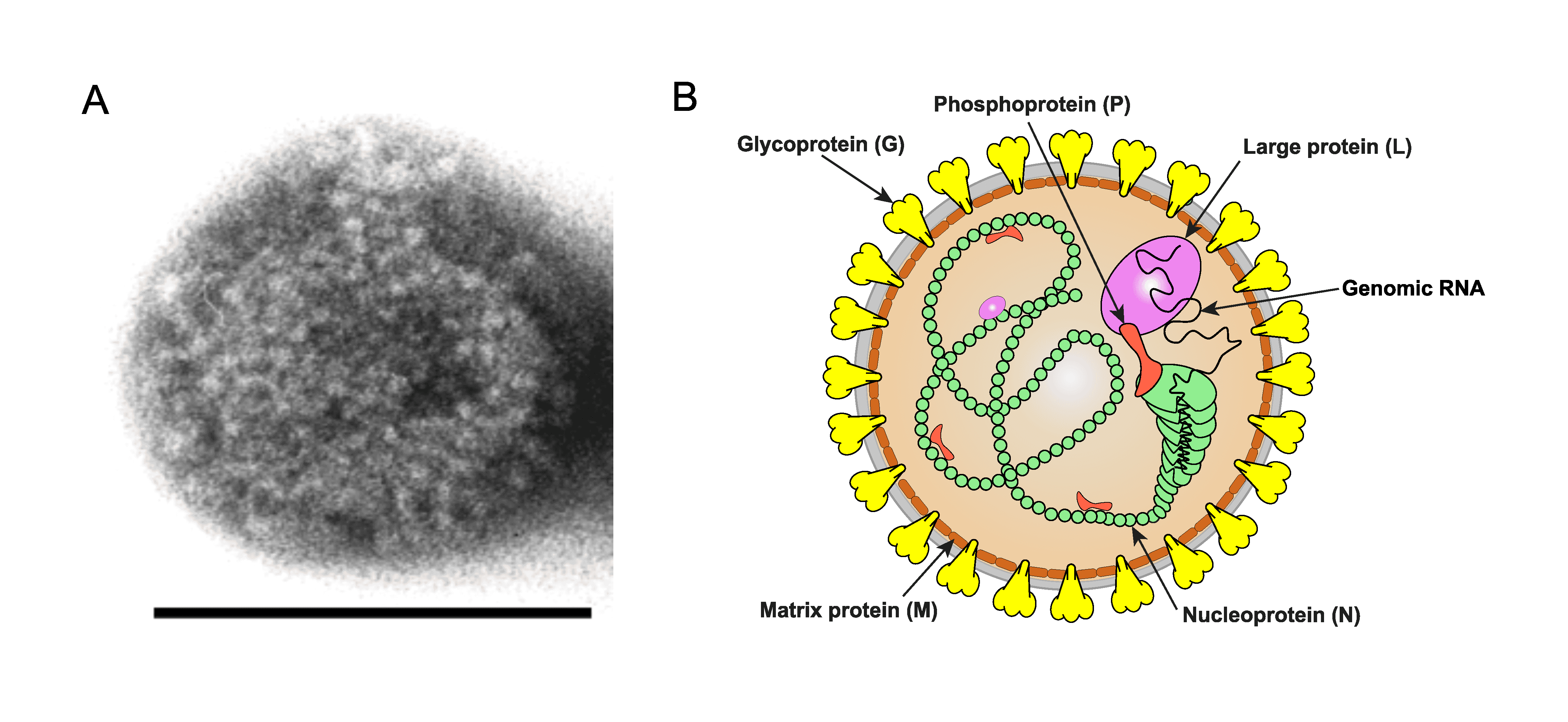 |
|
Figure 1.Orthobornavirus. Orthobornavirus particle structure. A) Electron micrograph of a Borna disease virus 1 particle. Size bar: 100 nm (Courtesy Dr M. Eickmann) B) Illustration of an orthobornavirus particle. Shown is the spherical enveloped particle with glycoprotein (G) spikes (yellow) inserted in a bilaminar lipid envelope (grey). |
Physicochemical and physical properties
See information provided in the family section.
Nucleic acid
The genome consists of a negative-sense non-segmented RNA of approximately 9 kb and a virion Mr of approximately 3×106. The RNA genome is not polyadenylated. Extracistronic sequences are found at the 3′- (leader) and 5′- (trailer) ends of the orthobornavirus genome. The ends of the orthobornavirus genome RNA exhibit partial inverted complementarity. Full-length positive-sense strand (antigenomic) RNAs are present in the nuclei of infected cells and in viral ribonucleoprotein (RNP) complexes. Defective RNAs have not been identified in Borna disease virus 1 (BoDV-1)-infected cells and tissues (Briese et al., 1992, Briese et al., 1994, Cubitt et al., 1994, Schneemann et al., 1995).
Proteins
The orthobornavirus virion is composed of the RNP, which consists of five structural proteins N, P, M, G, and L (Schwemmle et al., 1998, Kohno et al., 1999, Walker et al., 2000, Eickmann et al., 2005). For further details, see Table 1.Orthobornavirus.
Lipids
Lipids comprise the envelope bilayer of the virion, which is host-cell-derived; details on composition or subcellular origin are lacking.
Carbohydrates
See information provided in the family section.
Genome organization and replication
Orthobornavirus genomes contain three transcription initiation sites (S signals), and four transcription termination/polyadenylation sites (T signals), giving rise to four main transcription units (Figure 2. Orthobornavirus) (Briese et al., 1994). A putative fifth T signal (t6) is recognizable at nucleotide 4774 of BoDV-1 genomes but is absent in the other orthobornavirus genomes (Tomonaga et al., 2002). Orthobornaviruses lack the characteristic configuration of T signal/intergenic (IG) region/S signal commonly found at the gene boundaries of other negative-sense single-stranded RNA viruses. Instead, T/S transcription signals of orthobornavirus genomes frequently overlap (Figure 2. Orthobornavirus). Some primary transcripts are post-transcriptionally processed by the cellular RNA splicing machinery (Schneider et al., 1994). The viral mRNAs are polyadenylated, and their 5′-ends contain a blocking group. The 5′-terminus of genomic RNA is trimmed to be monophosphorylated and forms an incomplete inverted terminal repeat (Reuter et al., 2010, Martin et al., 2011).
 |
|
Figure 2. Orthobornavirus. Genome organization and transcription strategy of viruses in the genus Orthobornavirus. Dashed lines represent introns removed during alternative splicing of transcripts. N: nucleoprotein; X: accessory protein; P: phosphoprotein; M: matrix protein; G: glycoprotein; L: large protein containing an RNA-directed RNA polymerase domain. S: transcription initiation site; T: transcription termination site. |
Transcription unit 1 (S1 to T1) encodes N, which may be translated starting from two alternative start codons. Alternative splicing of this transcription unit has been identified for BoDV-1 and possibly Borna disease virus 2 (BoDV-2), resulting in additional isoforms of the N protein (Kojima et al., 2019).
Transcription unit 2 (S2 to T2) generates a bicistronic mRNA that encodes X and P. The X ORF starts upstream of the P ORF and overlaps, in a different frame with P. Orthobornavirus X proteins harbor a putative nuclear localization signal (NLS) in the N-terminus of the sequence. Although the biological role of X remains unclear, the X protein is necessary for viral replication and inhibits viral polymerase activity in polymerase reconstitution assays. A small ORF present upstream of the X/P gene affects the translation efficiency of X/P proteins. However, this ORF does not exist in all orthobornavirus genomes (Schneider et al., 1994, Watanabe et al., 2009, Fujino et al., 2012).
Transcription units 3 (S3 to T3) and 4 (S3 to T4) of orthobornaviruses have at least two introns (Figure 2. Orthobornavirus) (Schneider et al., 1994, Schneemann et al., 1995). By varying combinations of the splicing of these two introns, combined with leaky ribosomal scanning, the M, G, and L proteins are all expressed from the same transcription units. Some BoDV-1 isolates appear to possess a third intron to produce transcripts with the capacity to encode two additional proteins with predicted masses of 8.4 kDa (p8.4) and 165 kDa (p165), but production of such proteins has not been reported. G is post-translationally modified by N-glycosylation and is proteolytically cleaved by cellular subtilisin-like proteases, such as furin, resulting in the N-terminal and C-terminal cleavage products GP-N and GP-C, respectively (Richt et al., 1998) (Table 1.Orthobornavirus).
The replication cycle of orthobornaviruses is summarized in Figure 3.Orthobornavirus.
Table 1.Orthobornavirus. Location and function of orthobornavirus proteins.
|
Protein |
Location, mass, and function |
|
Nucleoprotein (N) |
A structural protein encoded by the first transcription unit. Encapsidates the viral RNA to form the viral nucleocapsid and together with L and P forms the ribonucleoprotein (RNP) complex (Briese et al., 1994, Cubitt et al., 1994, Schwemmle et al., 1998). Forms tightly assembled homotetramers of a cuboid-shaped structure that encloses a central channel (Rudolph et al., 2003). N is the target for immunopathologic responses of infected incidental mammalian hosts (Hausmann et al., 1999). Two major isoforms of 40 kDa and 38 kDa and additional less-abundant isoforms are synthesized (Kojima et al., 2019). The expression ratio of the isoforms contributes to the regulation of polymerase activity in virus replication (Schneider et al., 2004). The 40 kDa isoform contains both a nuclear localization signal (NLS, located in the N-terminus; P3KRRLVDDA11)* and a nuclear export signal (NES; L128TELEISSIFSHCC141)*, whereas the N-terminally truncated 38 kDa isoform possesses only an NES. |
|
Accessory protein (X) |
Multifunctional non-structural protein of 10 kDa encoded by the second transcription unit together with P (Poenisch et al., 2007). X modulates transcription and replication through binding to P as a regulator of viral transcriptase activity (Poenisch et al., 2004); essential for the viral replication cycle. Suppresses apoptosis in BoDV-1 infection (Poenisch et al., 2009). X has an NLS at its N-terminus (R6LTLLELVRRNGN19)*; the nuclear import of X is mediated by binding of its NLS to importin-α (Honda and Tomonaga 2013). |
|
Phosphoprotein (P) |
Structural protein of 24 kDa encoded by the second transcription unit and phosphorylated at serine residues (Briese et al., 1994). Component of the RNP (Briese et al., 1994, Cubitt et al., 1994, Schwemmle et al., 1998). Interacts with itself, N, X, M, and L (Schwemmle et al., 1998). BoDV-1 P is an acidic polypeptide (predicted isoelectric point of 4.8), that has a high Ser-Thr content (16%), with phosphorylation at serine residues that is mediated by both protein kinase C and casein kinase II (Schwemmle et al., 1997). These features are consistent with P acting as a transcriptional activator as found in other negative-sense single-stranded RNA viruses (Schwemmle et al., 1998). BoDV-1 P contains a bipartite NLS at the N-terminus (P29RPRKIPR36)* and C-terminus (P181PRIYPQLPSAPT193)*. It has a unique NES (M145KTMMETMKLMMEKVDLLYAS165)* within its methionine-rich domain. |
|
Matrix protein (M) |
Non-glycosylated, structural matrix protein of 14.5 kDa, encoded by the third transcription unit (Kraus et al., 2001); forms non-covalently linked tetrameric subunits associated with the inner surface of the viral membrane (Neumann et al., 2009). M is associated with viral RNPs in the host-cell nucleus (Chase et al., 2007, Hirai et al., 2016). |
|
Glycoprotein (G) |
Type I membrane surface glycoprotein and class III fusion protein (White et al., 2008, Garry and Garry 2009) generated from the G ORF of the third transcription unit after splicing of intron I (Briese et al., 1994, Cubitt et al., 1994). The primary translation product has a predicted mass of 56 kDa but is post-translationally modified to yield the glycosylated 84/94-kDa full-length structural protein (Eickmann et al., 2005). It is processed by cellular proteases into a C-terminal fragment of 43 kDa (GP-C or GP2) and an N-terminal fragment of 45–55 kDa (GP-N or GP1) (Richt et al., 1998). G and its derivatives are involved in orthobornavirion entry by attaching to an unknown receptor and mediating the fusion of endosomal and viral membranes (Gonzalez-Dunia et al., 1997, Gonzalez-Dunia et al., 1998, Perez et al., 2001). Antibodies to BoDV-1 G have neutralizing activity (Furrer et al., 2004). |
|
Large protein (L) |
This structural protein has a mass of 190 kDa, is encoded by a spliced ORF on the fourth transcription unit, and includes an RNA-directed RNA polymerase (RdRP) domain (Briese et al., 1994, Cubitt et al., 1994, Walker et al., 2000). L is homologous to the L proteins of other members of realm Riboviria. L is phosphorylated by cellular kinases, contains an NLS (R844VVKLRIAP852)*, interacts with P, and is a component of the viral RNP complex (Honda and Tomonaga 2013). |
* Examples for NLSs and NESs are provided for BoDV-1 (Honda and Tomonaga 2013).
 |
|
Figure 3.Orthobornavirus. Replication cycle of viruses of the genus Orthobornavirus (shown for Borna disease virus 1. (1) Virion uptake and entry: Attachment is mediated by binding of N-terminal cleavage product GP-N (Perez et al., 2001) to a yet uncharacterized cellular receptor (Duchala et al., 1989, Gonzalez-Dunia et al., 1998) by utilizing lipid rafts of binding immunoglobulin protein (BiP), a chaperone protein, in the plasma membrane (Honda et al., 2009). (2) Endocytosis: The virus is internalized by receptor-mediated endocytosis (Gonzalez-Dunia et al., 1998). (3) pH-dependent conformational change of the C-terminal cleavage product GP-C mediates the fusion of endosomal and viral membranes, resulting in release of ribonucleoproteins (RNPs) into the cytosol (Bajramovic et al., 2003): Nuclear import is regulated by nuclear localization signals (NLSs) and nuclear export signals (NESs) in the RNP components (Honda and Tomonaga 2013). (4) Nuclear replication results in positive-sense strand anti-genomic intermediates (complementary RNA [cRNA]) that serve as templates for viral genomic RNA (vRNA) (Briese et al., 1992). (5) Transcription and splicing: Four polyadenylated primary mRNA transcripts are transcribed from the vRNA, at least two of which are post-transcriptionally modified by cellular RNA splicing machinery to yield additional mRNA forms (Schneider et al., 1994, Tomonaga et al., 2002). (6) Translation: Viral mRNAs are translated after transport to the cytosol; translated RNP components (N, P, and L) are imported into the nucleus through their nuclear localization signals. (7) Virion assembly and budding: After intranuclear transport of their protein components, RNPs are assembled in the nucleus, and assembled RNPs are exported from the nucleus (Honda and Tomonaga 2013); after posttranslational cleavage of the G precursor protein, both resulting subunits (GP-N and GP-C) are transported to the cell surface to participate in the budding process (Richt et al., 1998); the non-glycosylated M protein is associated with the inner layer of the viral membrane (Kraus et al., 2001, Tomonaga et al., 2002). ER: endoplasmatic reticulum; RNP: ribonucleoprotein; N: nucleoprotein; X: accessory protein; P: phosphoprotein; M: matrix protein; G: glycoprotein; L: large protein. |
Biology
Host range
Members of the genus have a broad range of natural hosts, including mammals, birds, and reptiles. However, viruses appear to be adapted to particular hosts or a narrow range of hosts that act as their natural reservoirs (Dürrwald et al., 2016, Rubbenstroth et al., 2019).
Infection and pathogenicity
Orthobornaviruses can establish life-long persistence in their hosts. In some hosts, viruses are strictly neurotropic whereas in others, many organs may be infected. Characteristics of infection and pathogenesis are best understood for BoDV-1, variegated squirrel bornavirus 1 (VSBV-1), and members of the species Orthobornavirus alphapsittaciforme. BoDV-1 and VSBV-1 are broadly cell- and organ-tropic in their natural reservoirs (shrews and squirrels, respectively). Infectious virions are shed in urine and feces, but disease is not induced in reservoir hosts (Nobach et al., 2015, Schlottau et al., 2017). Rather, these viruses appear to be strictly neurotropic in accidental hosts (i.e., humans, horses, and other mammals) and can induce lethal encephalitis (Hoffmann et al., 2015, Dürrwald et al., 2016, Niller et al., 2020). The original reservoir hosts of members of the species Orthobornavirus alphapsittaciforme are so far unknown. However, they can infect a broad range of psittaciform birds (parrots) in captivity, with widespread cell and organ tropism and viral shedding. The clinical outcome in infected parrots is highly variable, ranging from inapparent to fatal neuropathic disease (Staeheli et al., 2010, Rubbenstroth et al., 2016). Orthobornaviruses are commonly not cytopathic, and they appear to induce disease mainly by T lymphocyte-mediated immunopathology in incidental hosts (Stitz et al., 2002). Two important pathogens in the veterinary sector are BoDV-1, which causes classical Borna disease in incidental hosts (e.g., horses and sheep), and psittaciform orthobornaviruses which cause proventricular dilatation disease (PDD) in parrots (Dürrwald et al., 2016). Additionally, BoDV-1 and VSBV-1 can cause fatal neuropathic disease in incidentally infected humans (Hoffmann et al., 2015, Niller et al., 2020).
Transmission
The natural transmission routes of orthobornaviruses are still largely unknown. Viral shedding by reservoir hosts occurs via various secretions and excretions and is usually correlated with virus being widely distributed in the organism. In contrast, incidental non-reservoir hosts with a strictly neurotropic infection are not known to shed detectable amounts of infectious virions. Horizontal transmission by an intranasal route has been confirmed for some orthobornaviruses in experimental infection models whereas, for others, the mode of transmission is unknown. Vertical transmission cannot be excluded, but firm evidence is missing (Dürrwald et al., 2016). Iatrogenic transmission of BoDV-1 by solid organ transplantation has been reported in Germany in three transplant recipients (Schlottau et al., 2018).
Geographical distribution
The geographical distribution of orthobornaviruses is linked to their respective reservoir hosts. Orthobornaviruses infecting hosts with a high mobility (migratory birds) or those that are extensively traded (exotic pet animals) have a wide distribution—possibly worldwide (Rubbenstroth et al., 2016). Others appear restricted to endemic areas due to more stationary reservoir hosts. For instance, BoDV-1, for which the bicolored shrew (Crocidura leucodon [Hermann, 1780]) is a reservoir, is confined to endemic areas in Germany, Austria, Switzerland, and Liechtenstein (Dürrwald et al., 2014, Rubbenstroth et al., 2019).
Antigenicity
Members of the genus Orthobornavirus share a broad serological cross-reactivity as demonstrated by indirect immunofluorescence assay, western blot, or enzyme-linked immunosorbent assay (ELISA) (Zimmermann et al., 2014). Antibodies in infected individuals are elicited primarily against the N and P antigens, but antibodies against X and M also have been detected. These antibodies are, however, not protective. Neutralizing antibodies directed against the G protein may be detected during prolonged persistent infection. Immunity and immunopathogenesis are mediated mainly by T lymphocytes. A contribution of neutralizing antibodies to immunity cannot be excluded (Stitz et al., 1998, Stitz et al., 2002).
Species demarcation criteria
Criteria for orthobornavirus species demarcation are primarily based on genomic characteristics, including PAirwise Sequence Comparison (PASC), in combination with biological characteristics, such as antigenic relationship and natural host range. In agreement with these additional criteria, the species differentiation cut-off for PASC of coding-complete genome sequences was defined as 72–75% (Kuhn et al., 2015).
Orthobornavirus sequences can be subdivided into four groups comprising (i) known mammalian bornaviruses and several avian bornaviruses (members of the species Orthobornavirus bornaense, Orthobornavirus sciuri, Orthobornavirus betapsittaciforme, Orthobornavirus serini, Orthobornavirus estrildidae and Orthobornavirus avisaquaticae) (ii) members of the species Orthobornavirus alphapsittaciforme, (iii) reptilian orthobornaviruses (members of the species Orthobornavirus elapsoideae and the unclassified Gaboon viper virus 1), and (iv) members of the species Orthobornavirus caenophidiae (Figure 4.Orthobornavirus).
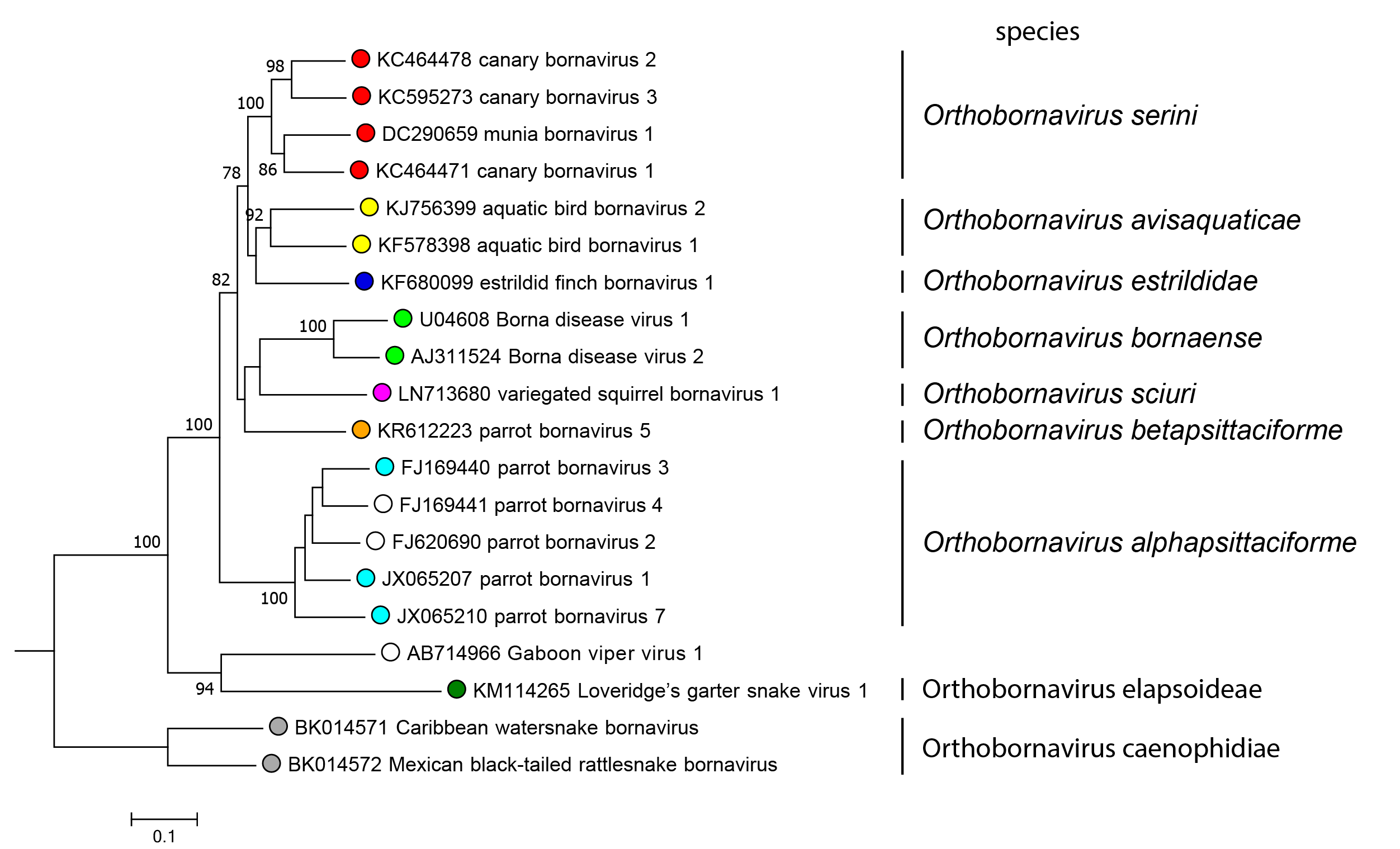 |
|
Figure 4.Orthobornavirus. Phylogeny of the genus Orthobornavirus. Complete P gene sequences (606 nucleotides) of representative orthobornaviruses were analyzed using the Neighbor-Joining algorithm and the Jukes-Cantor distance model in Geneious Prime bioinformatics software. Values at branches represent support in 1,000 bootstrap replicates. Only bootstrap values ≥70 at major branches are shown. Open circles indicate viruses that are unclassified due to insufficient sequence information. |
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
Gaboon viper virus 1 |
GaVV-1 |
Virus names and virus abbreviations are not official ICTV designations.

