Family: Alphaflexiviridae
Genus: Allexivirus
Distinguishing features
Allexiviruses are distinguished by mite transmission and by the presence of a large conserved ORF4 or 5 after the position where the third, and smallest, of the triple gene block (TGB) proteins, TGB3, is found in the other plant-infecting members of the family. Whereas the start codon is missing for the TGB3 ORF in most (but not all) genus members, the coding capacity is conserved and the ORF may be translated through alternative mechanisms.
Virion
Morphology
Virions are highly flexible filamentous particles, about 800 nm in length and 12 nm in diameter. They resemble potyviruses in their length, but closteroviruses in their flexibility and cross-banded substructure (Figure 1. .Alphaflexiviridae).
Physicochemical and physical properties
Virions of shallot virus X sediment with an S20,w of about 170S in 0.1 M Tris-HC1, pH 7.5 at 20°C and have a buoyant density in CsCl of 1.33 g cm−3.
Nucleic acid
Virions contain a single molecule of linear single-stranded RNA, of about 9.0 kb, with a 3′-poly(A) tract. The genomic RNA contains six large ORFs and short untranslated regions (UTR) at the 5′- and 3′-termini (Figure 2. .Alphaflexiviridae).
Proteins
Virions are composed of a 25–32 kDa polypeptide as a major capsid protein (CP). A 42 kDa polypeptide is a minor component of virions.
Carbohydrates
None reported.
Genome organization and replication
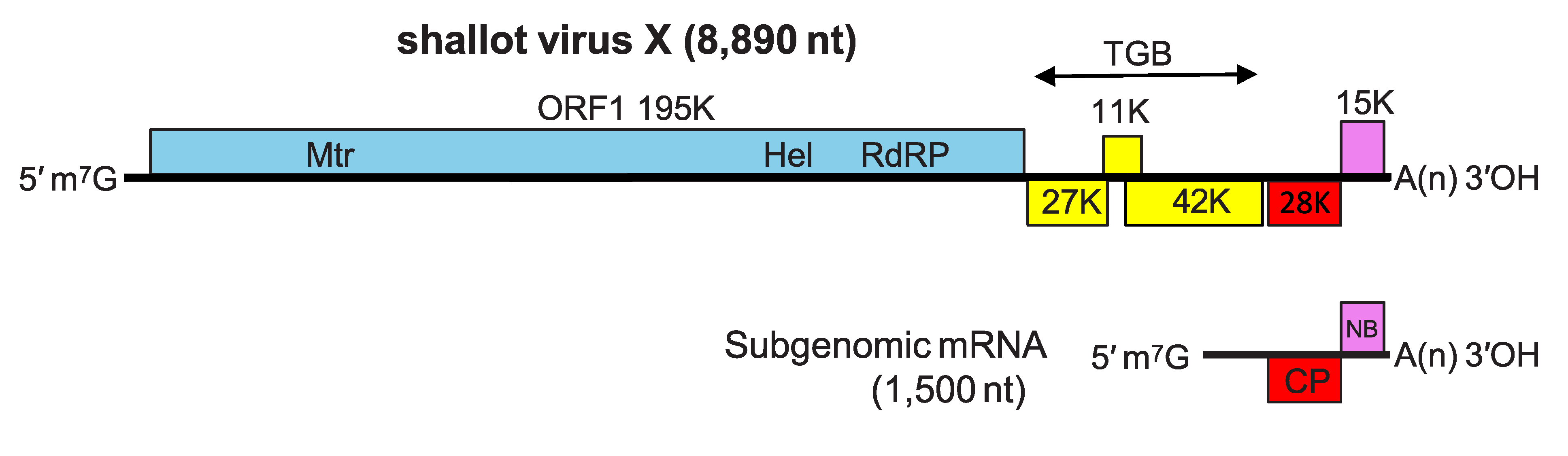
The genomic RNA contains six large ORFs and short UTRs at the 5′- and 3′-termini (Kanyuka et al., 1992) (Figure 1. Allexivirus) The ORFs code for polypeptides of about 195, 27, 11, 42, 28 and 15 kDa, respectively from the 5′-end to the 3′-end. The 195 kDa polypeptide is the replication-associated (Rep) protein. The 26 and 11 kDa proteins are similar to the first two proteins encoded by the TGB of related plant viruses and are probably involved in cell-to-cell movement of the virus. There is a coding sequence for a third small (7–8 kDa) TGB protein but the initiation AUG-codon is lacking in most, but not all member species. The 42 kDa polypeptide (ORF4) has no significant homology with any known protein; in plants infected with an isolate of the type species, the 42 kDa protein was expressed in relatively large amounts and was shown to be involved in virion assembly. The 28 kDa polypeptide is the CP. In SDS-polyacrylamide gel electrophoresis it migrates as an apparently 32–36 kDa protein, which could be due to its high hydrophilicity. The 15 kDa protein has a zinc-binding finger motif and an ability to bind nucleic acids. The function of this polypeptide is not known.
 |
|
Figure 1. Allexivirus. Genome organization and translation strategy of shallot virus X showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; Hel, helicase; RdRP, RNA-directed RNA polymerase; TGB, triple gene block; CP, capsid protein; NB, nucleic acid binding protein. |
Biology
Host range
The host range of allexiviruses that encode a 15 kDa RNA-binding protein is mainly restricted to Allium species. Some isolates from shallot, onion, garlic and sand leek have been experimentally transmitted to Chenopodium murale, in which they induced local lesions. Viruses lacking the RNA-binding protein infect hosts other than Allium including vanilla, alfalfa, groundnut and blackberry.
Transmission
Allexiviruses are thought to be mite-borne. Garlic viruses C and D have been shown to be transmitted by the eriophyd mite, Aceria tulipae. All are manually transmissible by sap inoculation to healthy host plants. None could be transmitted by aphids.
Geographical distribution
Allexiviruses are widely distributed and probably occur wherever their host plants are grown.
Cytopathic effects
Most allexiviruses induce no visible or only very mild symptoms, although certain isolates can cause severe damage to crops. In infected tissue, allexiviruses can induce the formation of granular inclusion bodies and small bundles of flexible particles.
Antigenicity
Allexivirus particles are immunogenic. Some members of the genus are serologically related. Specific antisera and monoclonal antibodies against virus particles as well as antisera against recombinant CPs have been used for differentiation purposes.
Derivation of names
Allexi: from Allium (the genus name for the principal host, shallot) and exi, a phonetic version of “X”.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Members of different species have less than 72% nt identity (or 80% aa identity) in their CP or Rep genes.
- Members of different species react differently with antisera.
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
allexivirus DS-2013/CZE |
|
|
|
Cassia mild mosaic virus |
|
|
|
Senna severe yellow mosaic virus |
|
|
|
shallot mite-borne latent virus |
|
Virus names and virus abbreviations are not official ICTV designations.

