Family: Chaseviridae
Hany Anany, Padmanabhan Mahadevan, Dann Turner, Evelien M. Adriaenssens and Andrew M. Kropinski
The citation for this ICTV Report chapter is the summary published as Anany et al., (2022):
ICTV Virus Taxonomy Profile: Chaseviridae 2022, Journal of General Virology 2022;103:001715
Corresponding author: Hany Anany ([email protected], [email protected])
Edited by: Evelien M. Adriaenssens and Stuart G. Siddell
Posted: November 2021, updated May 2023, July 2025
Summary
Members of the family Chaseviridae are bacterial viruses infecting representatives of the bacterial class Gammaproteobacteria. Virions of members of this family have myovirus morphology, i.e., a head-tail structure with a long, contractile tail, and an icosahedral head (Table 1 Chaseviridae). Genomes are dsDNA of 52–56 kbp with G+C content ranging from 39.3–52.5%. Chaseviruses, like members of the family Autographiviridae, encode a large single subunit RNA polymerase, but unlike those viruses their promoter sequences have not yet been identified.
Table 1 Chaseviridae Characteristics of members of the family Chaseviridae
| Characteristic | Description |
| Example | Escherichia phage vB_EcoM-4HA13 (MN136198), species Sabourvirus sv4HA13, genus Sabourvirus |
| Virion | Head-tail morphology with contractile tail; heads generally isometric with diameters of 53–65 nm showing capsomers, uncontracted tails of 116–166 nm in length |
| Genome | Linear, terminally redundant, non-permuted dsDNA of 52–56 kbp |
| Replication | Phage-encoded DNA polymerase |
| Translation | Bacterial translation |
| Host range | Bacteria of the phylum Proteobacteria, class Gammaproteobacteria |
| Taxonomy | Realm Duplodnaviria, kingdom Heunggongvirae, phylum Uroviricota, class Caudoviricetes; 2 subfamilies (Cleopatravirinae, Nefertitivirinae), 13 genera and 30 species |
Virion
Morphology
Virions have isometric, icosahedral heads of 53–65 nm in diameter. The heads show clear capsomers, i.e. the subunits of the capsid are arranged in pentons and hexons that are assembled into the isometric, icosahedral capsid. The contracted tails are 116–166 nm in length. (Figure 1 Chaseviridae).
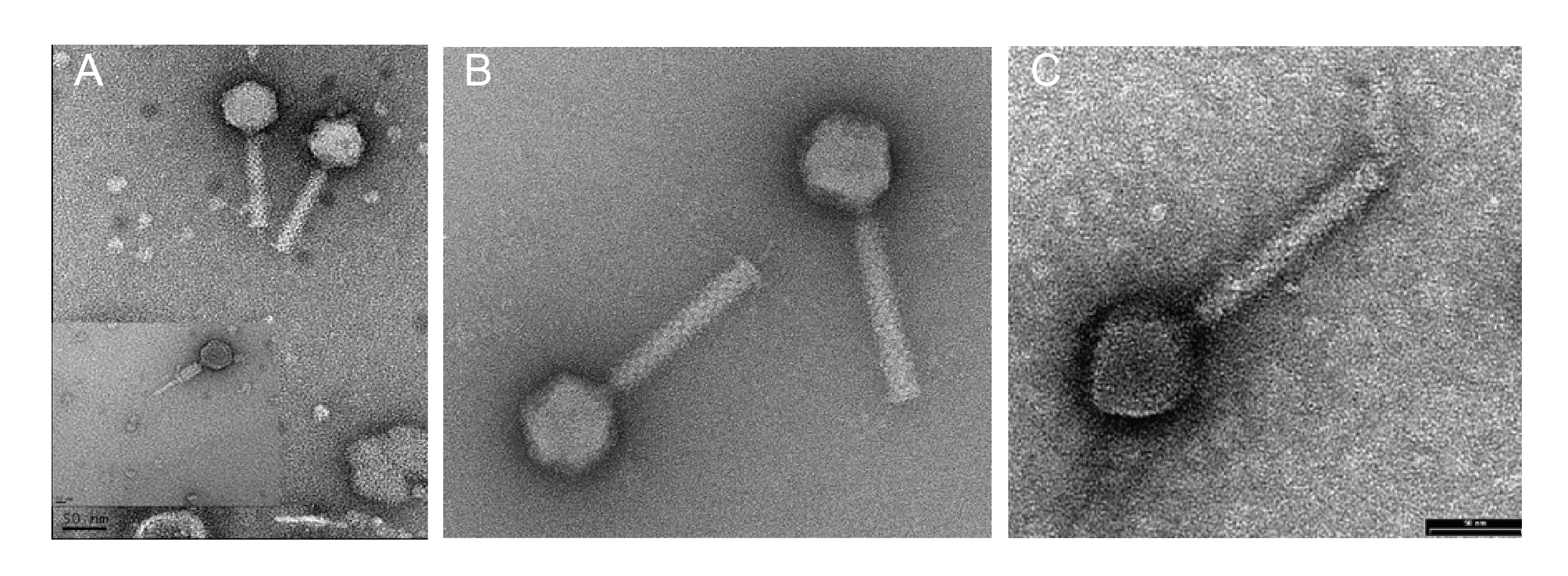 |
| Figure 1 Chaseviridae. Transmission electron micrographs of negatively stained phages. (A) Escherichia phage vB_EcoM-4HA13, (B) Erwinia phage vB_EamM-Y2 (provided by Martin J. Loessner) and (C) Escherichia phage phiEcoM-GJ1 (provided by Nidham Jamalludeen). |
Physicochemical and physical properties
Not determined.
Nucleic acid
The genomes of members of the family Chaseviridae consist of linear dsDNA with long terminal repeats of approximately 3 kbp. Genomes range between 52–56 kbp. Some members are predicted to encode tRNAs. There is no report of modified bases in the DNA of these viruses. A genome diagram of Escherichia phage vB_EcoM-4HA13, is typical of the family (Figure 2 Chaseviridae).
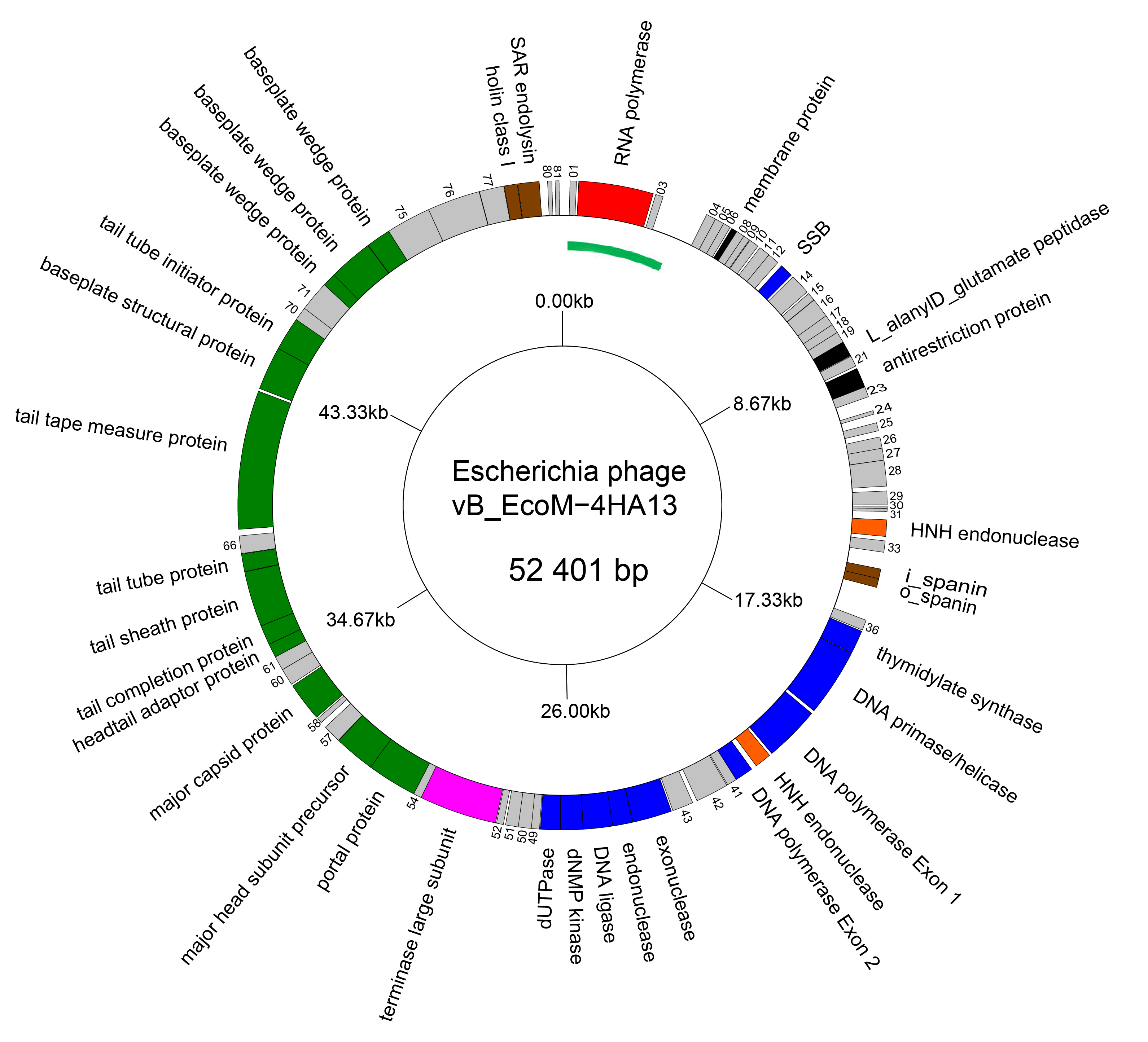 |
| Figure 2 Chaseviridae Genome organisation of Escherichia phage vB_EcoM-4HA13. Genes involved in transcription are coloured red, DNA and nucleotide synthesis, blue; homing endonucleases, orange; packaging, pink; morphogenesis, green; and lysis brown. The linear genome is represented as a circle with the green arc representing the left LTR. This figure was generated with GenomeVx (Conant and Wolfe 2008) and edited with Foxit PDF Editor. |
Proteins
The virion proteins of three members of this family, Escherichia phage phiEcoM-GJ1 (Jamalludeen et al., 2008), Escherichia phage vB_EcoM-4HA13 and, Erwinia phage vB_EraM-Y2 (Born et al., 2011) have been investigated using mass spectrometry. In the case of Escherichia phage vB_EcoM-4HA13, 15 structural proteins were identified with the major capsid protein containing an acetylated lysine residue.
Genome organization and replication
The genomes of chaseviruses are linear dsDNA with long terminal repeats of approximately 3 kbp. Genomes are of 52–56 kbp and encode 63–92 genes, including 0–1 tRNAs. For Escherichia phage vB_EcoM-4HA13, all coding sequences are transcribed off a single strand (Figure 2. Chaseviridae). Transcription is mediated by the host and a phage-encoded Autographiviridae-like RNA polymerase. DNA replication is mediated by a phage-encoded DNA polymerase and DNA helicase/primase.
Comparing all chaseviruses at the protein level, using CoreGenes 5.0 (https://coregenes.ngrok.io/ and by ViPTree analysis (Nishimura et al. 2017)) with the OrthoMCL algorithm (Li et al., 2003), reveals 25 conserved proteins representing 33.7% of the viral proteome. The conserved proteins were identified using PHROGS ((Terzian et al., 2021); https://phrogs.lmge.uca.fr/) and are presented in Table 2 Chaseviridae.
Table 2 Chaseviridae. Core genes (with predicted functions) shared among all members of the family (identified using CoreGenes 5.0 and mapped to the PHROGS database).
| NCBI prot ID* | PHROG number | PHROG annotation | PHROG category |
| YP_001595396.1 | phrog_414 | RNA polymerase | DNA, RNA and nucleotide metabolism |
| YP_001595423.1 | phrog_13087 | unknown function | unknown function |
| YP_001595430.1 | phrog_239 | DNA primase/helicase | DNA, RNA and nucleotide metabolism |
| YP_001595432.1 | phrog_17 | DNA polymerase | DNA, RNA and nucleotide metabolism |
| YP_001595434.1 | phrog_862 | unknown function | unknown function |
| YP_001595435.1 | phrog_107 | RNaseH | DNA, RNA and nucleotide metabolism |
| YP_001595436.1 | phrog_1985 | HNH endonuclease | DNA, RNA and nucleotide metabolism |
| YP_001595438.1 | phrog_139 | deoxynucleoside monophosphate kinase | other (intermediary metabolism) |
| YP_001595443.1 | phrog_38 | terminase large subunit | head and packaging |
| YP_001595445.1 | phrog_146 | portal protein | head and packaging |
| YP_001595446.1 | phrog_530 | head maturation protease | head and packaging |
| YP_001595447.1 | phrog_563 | virion structural protein | head and packaging |
| YP_001595448.1 | phrog_264 | major head protein | head and packaging |
| YP_001595450.1 | phrog_302 | virion structural protein | head and packaging |
| YP_001595451.1 | phrog_11887 | head protein | head and packaging |
| YP_001595452.1 | phrog_12771 | tail completion | connector |
| YP_001595453.1 | phrog_205 | virion structural protein | head and packaging |
| YP_001595454.1 | phrog_280 | virion structural protein | head and packaging |
| YP_001595455.1 | phrog_11820 | tail assembly chaperone | tail |
| YP_001595458.1 | phrog_196 | virion structural protein | head and packaging |
| YP_001595461.1 | phrog_11 | baseplate protein | tail |
| YP_001595462.1 | phrog_120 | baseplate protein | tail |
| YP_001595463.1 | phrog_6 | baseplate protein | tail |
| YP_001595464.1 | phrog_587 | structural protein | head and packaging |
* Escherichia phage phiEcoM-GJ1 (Jamalludeen et al., 2008)
Biology
Viruses in the family Chaseviridae are obligately lytic and infect bacteria belonging to the phylum Proteobacteria, class Gammaproteobacteria and have a worldwide distribution.
Derivation of names
Cleopatravirinae: from Cleopatra VII Philopator (69 – 30 BCE) who was the last active ruler of the Ptolemaic Kingdom in Egypt.
Carltongylesvirus: named in honour of Dr. Carlton L. Gyles (1940 – 2025) in whose laboratory at the University of Guelph, Ontario, Canada, Escherichia phage phiEcoM-GJ1 was isolated and studied.
Chaseviridae – The family name honours Martha Cowles Chase (1927 – 2003), also known as Martha C. Epstein, an American geneticist known for experimentally confirming (with Alfred Hershey) that DNA rather than protein is the genetic material of life (https://en.wikipedia.org/wiki/Martha_Chase).
Faunusvirus: from the name of the exemplar isolate of the species Faunusvirus faunus, Erwinia phage Faunus.
Fifivirus: from the names of two of the phages is this taxon, Erwinia phages Fifi44 and Fifi451
Liaoningvirus: from Liaoning Province (China) where in the School of Bioengineering, Dalian University of Technology the first isolate of its type, Vibrio phage vB_VpaM_VPs20, was isolated
Loessnervirus: named in honour of Professor Dr. Martin J. Loessner, ETH Zurich, Switzerland, in whose laboratory Erwinia phage vB_EamM-Y2 was first isolated and studied.
Longwangvirus: from Chinese mythology, where Longwang (龍王) lords over the seas and is known as the “Dragon King.”
Myducvirus: from the first virus of this genus, Proteus phage Myduc.
Nefertitivirinae – from Neferneferuaten Nefertiti (c. 1370 – c. 1330 BCE) who was an Egyptian queen and the Great Royal Wife of Akhenaten, an Egyptian Pharaoh.
Pahsextavirus: from the first isolate of a virus from this genus, Aeromonas phage pAh6-C, with the number six replaced with the Latin prefix sexta.
Phayathaivirus: derived from the address (254 Phayathai, Bangkok, Thailand), where at Chulalongkorn University the first of these viruses was isolated.
Sabourvirus: named in honour of Dr. Parviz M. Sabour (1936 – 2025) who was the first Agriculture and Agri-Food Canada scientist to carry out research on the agricultural application of bacteriophages.
Shantouvirus: This genus is named after Shantou, a city in southern China.
Suwonvirus: This genus is named after Suwon, the capital of Gyeonggi Province, in north-western South Korea where Pectobacterium phage PM1 was first isolated.
Yushanvirus: from Yushan Road, the location of the Ocean University of China, College of Food Science & Engineering where Shewanella phage Spp001 was isolated.
Subfamily demarcation criteria
Subfamilies (Table 3 Chaseviridae) are identified as well-supported monophyletic groups based on phylogenetic analysis of the RNA polymerase and terminase large subunit proteins (Figure 3 Chaseviridae). Within each subfamily, members are 25–29% similar in their whole genome sequence as calculated with the VIRIDIC algorithm.
Table 3 Chaseviridae. The subfamilies of Chaseviridae
| Subfamily | Genera | Hosts | Genome (kbp)* | Terminal repeats |
| Cleopatravirinae | 6 | Erwinia sp, Escherichia sp, Pantoea sp, Pectobacterium sp, Proteus sp | 53.45 | ND** |
| Nefertitivirinae | 2 | Aeromonas sp, Shewanella sp, Vibrio sp. | 52.48
| ca. 3 kbp |
*Average; **ND Not determined for any member
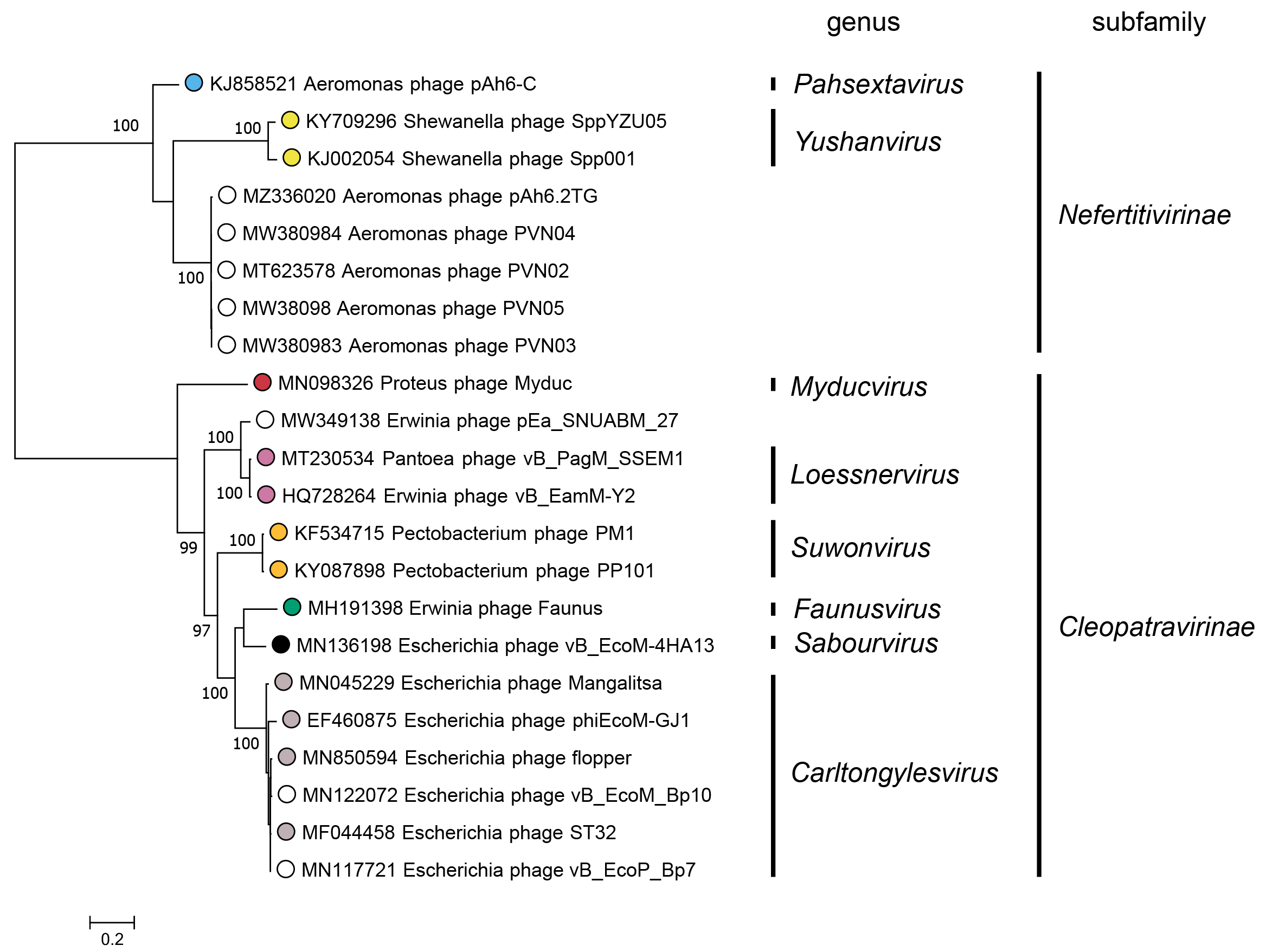 |
| Figure 3 Chaseviridae. Partitioned Maximum Likelihood phylogenetic tree of the RNA polymerase and terminase large subunit amino acid sequences of members of the family Chaseviridae. For each predicted protein, amino acid sequences were aligned with MAFFT (Katoh and Standley 2013), and trimmed with Trimal using the setting “gappyout” (Capella-Gutiérrez et al., 2009). The phylogenetic tree was constructed using the IQ-Tree suite, using the partitioned analysis with mixed data setting using as input the full multiple sequence alignment of each protein sequence with ultrafast bootstraps (UFBoot) calculated on 1000 repetitions (Nguyen et al., 2015, Chernomor et al., 2016, Hoang et al., 2018). For both genes, the best fitting substitution model was LG+G4 as determined using ModelFinder on each alignment separately, based on the Bayesian Information Criterion (Kalyaanamoorthy et al., 2017). The log-likelihood of the final tree was -18292. The consensus tree was drawn midpoint rooted with unclassified viruses indicated by open circles. UFBoot values above 95% are shown at nodes. This phylogenetic tree and corresponding sequence alignment files are available to download from the Resources page. |
Genus demarcation criteria
Genera in the family Chaseviridae are well-supported monophyletic clades by VIRIDIC clustering and in (concatenated) marker gene phylogenies. Members of a genus share at least 70% nucleotide identity across the genome. Members of the same genus generally infect members of the same bacterial genu
Species demarcation criteria
Members of the same species are more than 95% identical in genome nucleotide sequence, including the terminal repeat region. Viruses with genomes that differ by more than 5% are assigned to different species.
Relationships within the family
A Maximum Likelihood tree for the RNA polymerase and terminase large subunit proteins (Figure 3 Chaseviridae) shows a clear separation between members of the two subfamilies Nefertitivirinae and Cleopatravirinae. Members of different genera are robustly separated into different clades. Within a genus, differences between members of different species are separated into reproducible clades as evidenced by short branch lengths and low bootstrap support. Nucleotide similarity over the entire genome length for species demarcation.
Relationships with other taxa
Viruses in the family Chaseviridae share morphological similarity with other myoviruses, i.e. dsDNA bacteriophages with contractile tails. A ViPTree which employs TBLASTx analyses shows deep branching of members of this family into two clades (Figure 4 Chaseviridae).
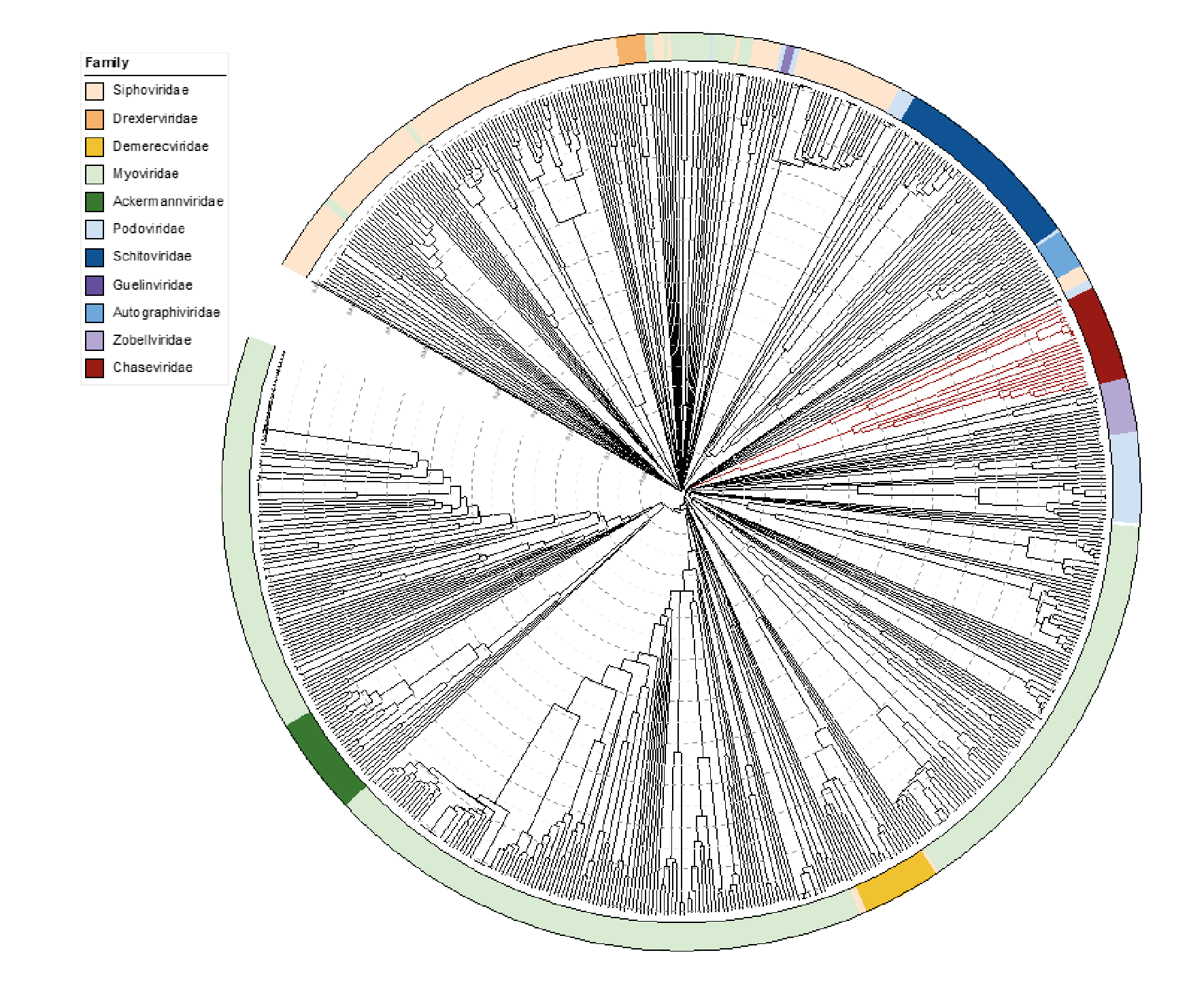 |
| Figure 4 Chaseviridae. ViPTree analysis (https://www.genome.jp/viptree/; (Nishimura et al., 2017)) is based upon the Phage Proteomic Tree (Rohwer and Edwards 2002). The tree was annotated in iTol (Letunic and Bork 2019) to display the virus families as a colour strip. Members of the Chaseviridae are indicated in dark red. The tree scale as indicated with concentric circles represents the similarity between genomes at the predicted proteome level on a scale from 0 to 1 (branch length 0 = identical, 1 = no detectable similarity). This phylogenetic tree is available to download from the Resources page. |
Related unclassified viruses
| Virus name | Accession No. | Reference |
| Erwinia phage pEa_SNUABM_27 | MW349138 | |
| Erwinia phage Fifi44 | OK073976 | |
| Escherichia phage vB_EcoP_Bp7 | MN117721 | |
| Escherichia phage vB_EcoM_Bp10 | MN122072 | |
| Escherichia phage vB_EcoM_DE15 | OP595144 | |
| Escherichia phage vB_EcoM_SA91KD | OL960574 | |
| Escherichia phage SKA49 | OL741059 | |
| Escherichia phage MLP1 | OK148439 | |
| Escherichia phage PO103-1 | ON778463 | |
| Aeromonas phage pAh6.2TG | MZ336020 | |
| Aeromonas phage PVN02 | MT623578 | |
| Aeromonas phage PVN03 | MW380983 | |
| Aeromonas phage PVN04 | MW380984 | |
| Aeromonas phage PVN05 | MW380985 | |
| Aeromonas phage PAS-1 | JF342690, JF342689, JF342688, JF342687, JF342686, JF3426841, JF342683(*) | (Kim et al., 2012) |
| Aeromonas phage PVN03 | MW380983 | |
| Aeromonas phage BUCT696 | OL770365 | |
| Vibrio phage vB_VpaM_VPs20 | OP056089 | |
| Yersinia phage PYps23T | MW147598 | |
| Yersinia phage PYps16N | MW147601 |
Virus names and virus abbreviations are not official ICTV designations.
(*) partial genome

