Family: Botourmiaviridae
María A. Ayllón, Massimo Turina, Jiatao Xie, Luca Nerva, Shin-Yi Lee Marzano, Livia Donaire and Daohong Jiang
The citation for this ICTV Report chapter is the summary published as Donaire et al., (2024):
ICTV Virus Taxonomy Profile: Botourmiaviridae 2024, Journal of General Virology, 105: 002047
Corresponding author: María A. Ayllón ([email protected])
Edited by: Sead Sabanadzovic
Posted: February 2020, updated September 2021, September 2022, June 2023, May 2024
Summary
The family Botourmiaviridae includes viruses with positive-sense RNA genomes infecting plants and fungi (Table 1 Botourmiaviridae). Some viruses of the family have been identified in leaves showing symptoms of infection with the oomycete Plasmopara viticola, suggesting that oomycetes could also be hosts of these viruses. The family includes twelve genera: Ourmiavirus, Botoulivirus, Betabotoulivirus, Magoulivirus, Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus, Penoulivirus, Rhizoulivirus and Betarhizoulivirus. Members of the genus Ourmiavirus are plant viruses with non-enveloped bacilliform virions composed of a single coat protein. The genome consists of three segments, each one encoding a single protein: RNA-directed RNA polymerase (RdRP), movement protein (MP) or coat protein (CP). Virions possess a unique structure with a series of discrete lengths from 30 to 62 nm. The RdRP has closest similarity to that of mitoviruses and narnaviruses; the MP is similar to that of tombusviruses; the CP shows limited similarity to the coat proteins of several plant and animal viruses. In general, members of the other genera are non-encapsidated fungal viruses with a monopartite and monocistronic genome of 2 000–5 300 nucleotides encoding an RdRP.
Table 1 Botourmiaviridae. Characteristics of members of family Botourmiaviridae
| Characteristic | Description |
| Example | Ourmia melon virus VE9 (RNA1: EU770623; RNA2:EU770624; RNA3: EU770625), species Ourmiavirus cucurbitae |
| Virion | Ourmiavirus: bacilliform (18 nm × 30–62 nm) with a 23.8 kDa coat protein. Members of other genera are not encapsidated |
| Genome | Positive-sense RNA; monopartite (Botoulivirus, Betabotoulivirus, Magoulivirus, Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus, Penoulivirus, Rhizoulivirus, and Betarhizoulivirus) (2–5 kb) or tripartite (Ourmiavirus) (2.8 kb; 1.1 kb; 0.97 kb) |
| Replication | Cytoplasmic; virion assembly is coupled to active replication |
| Translation | From genomic RNA; each genomic segment is monocistronic |
| Host Range | Plants and fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Lenarviricota, class Miaviricetes, order Ourlivirales; the family Botourmiaviridae includes 12 genera (Ourmiavirus, Botoulivirus, Betabotoulivirus, Magoulivirus, Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus, Penoulivirus, Rhizoulivirus and Betarhizoulivirus) each with multiple species, for a total of 159 species |
Virion
Morphology
The bacilliform virions of ourmiaviruses constitute a series of particles with conical ends (apparently hemi-icosahedra) and cylindrical bodies 18 nm in diameter. The bodies of the particles are composed of a series of double disks, the most common particles having two disks (particle length 30 nm), a second common particles composed of three disks (particle length 37 nm) with rarer particles built-up of four (particle length 45.5 nm) or six disks (particle length 62 nm) (Figure 1 Botourmiaviridae). There is no envelope (Figure 2 Botourmiaviridae).
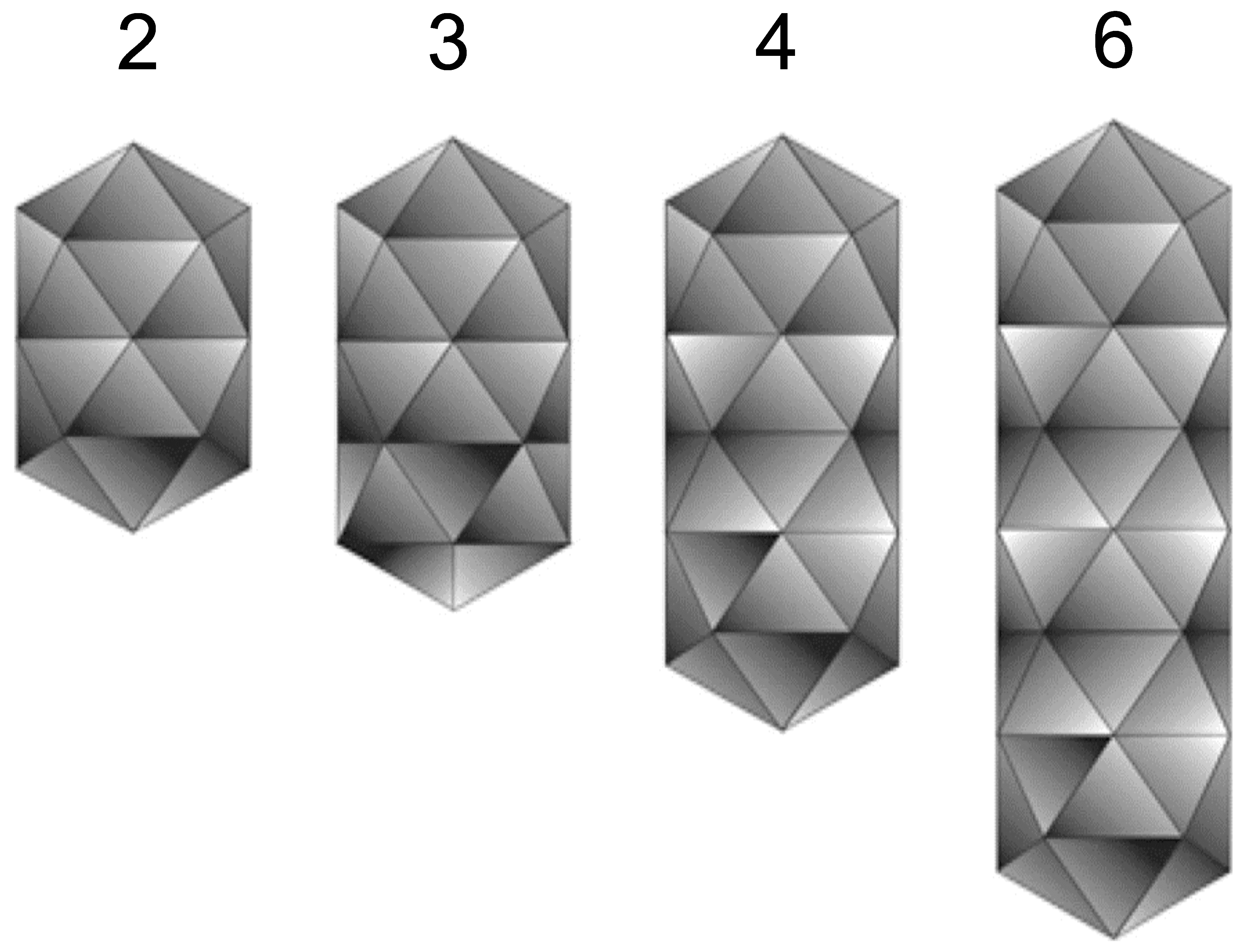 |
| Figure 1 Botourmiaviridae. Diagram of virion surface of a member of the genus Ourmiavirus, showing arrangement and number of double disks and conical ends in particles of different length. Each row of five triangles represents a double disk. |
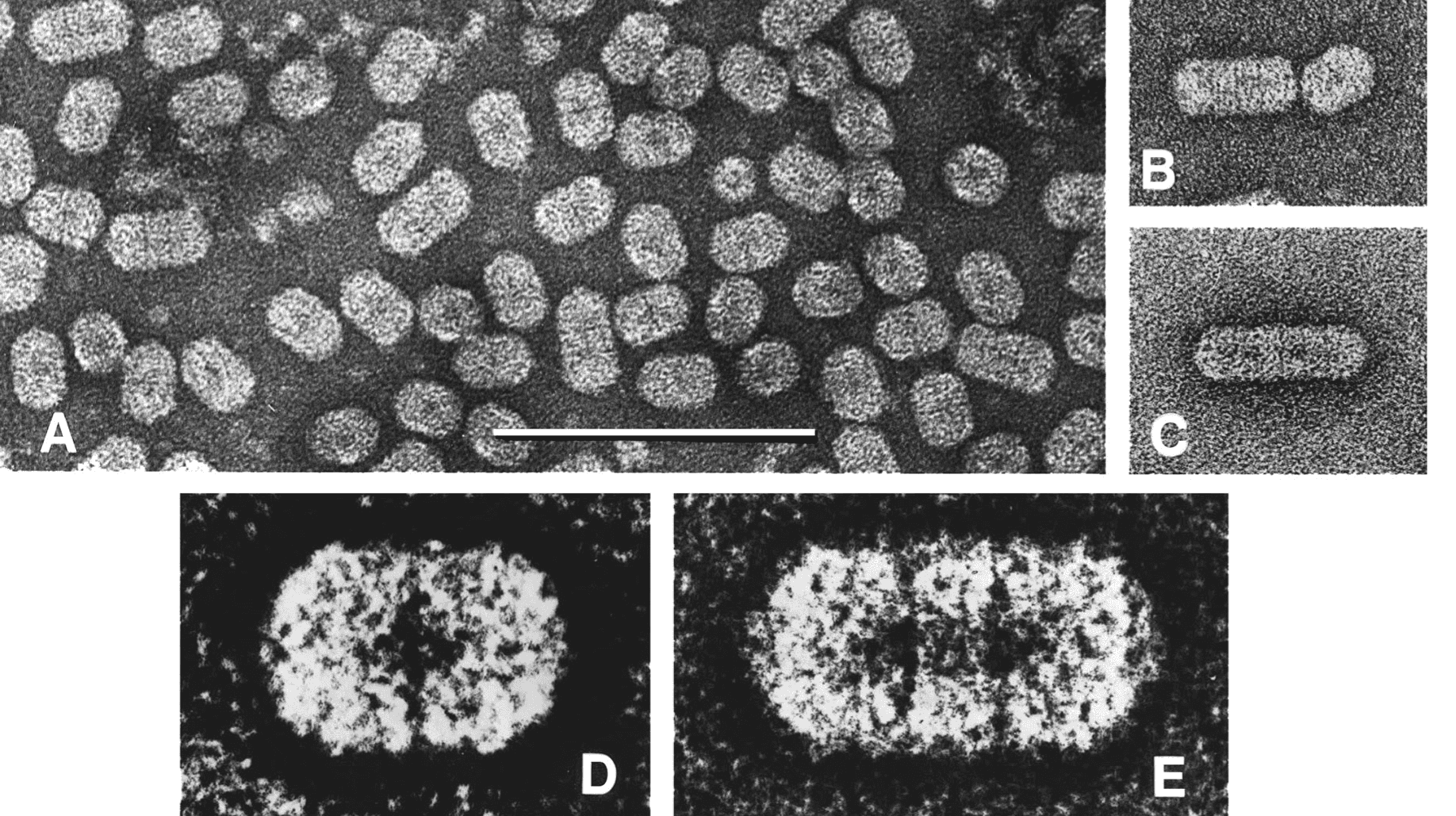 |
| Figure 2 Botourmiaviridae. Virion morphology. (A, B, C) Negative-contrast electron micrographs (uranyl acetate) of purified particles of Ourmia melon virus. The bar represents 100 nm. (D, E) Features of the two commonest particle types, enhanced by photographic superimposition. |
Physicochemical and physical properties
For members of the genus Ourmiavirus the Mr of virions and their sedimentation coefficients are not known. The buoyant density in CsCl of all particle sizes is 1.375 g cm−3. The particles are stable at pH 7. Ourmiaviruses are relatively thermally stable and retain infectivity in crude sap after heating for 10 min at 70 °C but not 80 °C, and after at least one freeze–thaw cycle. The particles are stable after CsCl density gradient centrifugation, treatment with Triton X-100, and treatment with chloroform but not n-butanol.
Nucleic acid
Ourmiaviruses have a genome comprised of three linear, positive-sense RNA molecules. Members of the other eleven genera have a monosegmented genome. In Ourmia melon virus (OuMV), the three RNAs are 2 814, 1 064 and 974 nt in size, similar to those of other members of the genus (Rastgou et al., 2009). In Botrytis ourmia-like virus (BOLV) (genus Botoulivirus) the RNA is 2 903 nt (Donaire et al., 2016); members of the genera Botoulivirus, Betabotoulivirus, Magoulivirus, Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus, Penoulivirus, Rhizoulivirus, and Betarhizoulivirus have genomes of a similar length. The genomes of Magnaporthe oryzae ourmia-like virus 1 isolate Guy11, a representative isolate of the genus Magoulivirus, the scleroulivirus Magnaporthe oryzae botourmiavirus 9 isolate SH05, and the gammascleroulivirus Magnaporthe oryzae botourmiavirus 6 are polyadenylated at the 3′-end (Illana et al., 2017); this is not observed for members of other genera.
Proteins
CP, the single structural protein of Ourmia melon virus is 23.8 kDa and is encoded by RNA3.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
In members of the family Botourmiaviridae, each genomic RNA has one open reading frame (ORF) (Figure 3 Botourmiaviridae). RNA1 of members of the genus Ourmiavirus, or the unique genomic RNA of viruses in the other genera, encode a protein containing the highly conserved core domain GDD typical of an RdRP (Rastgou et al., 2009, Donaire et al., 2016, Marzano and Domier 2016, Marzano et al., 2016, Illana et al., 2017). Ourmia melon virus encodes a 23.8 kDa CP (encoded by RNA3) and the two non-structural proteins RdRP (97.5 kDa, encoded by RNA1) and the MP (31.6 kDa, encoded by RNA2) (Crivelli et al., 2011). Similarly-sized proteins are predicted to be encoded by members of the species, Epirus cherry virus and cassava virus C (Rastgou et al., 2009). For botourmiaviruses infecting fungi, the single ssRNA segment encoding RdRP is sufficient for replication (Wang et al., 2020).
 |
| Figure 3 Botourmiaviridae. Schematic genome organization of representative isolates of the twelve genera of the family Botourmiaviridae showing the size of each RNA and the positions and coding capacity of the ORFs. CP, coat protein; MP, movement protein; RdRP, RNA-directed RNA Polymerase. |
A protein fusion of the CP to GFP localizes specifically to the nucleolus (Rossi et al., 2014) but there is no direct evidence of the CP in the nucleus during infection (Rossi et al., 2015). Synthesis of CP from actively replicating RNA3 is necessary for both virion assembly and systemic infection of the host (Crivelli et al., 2011). The MP may undergo post-translational modification. Alanine scanning mutagenesis of conserved residues in the MP showed their importance in determining symptoms, movement, and formation of tubular structures that may play a role in cell-to-cell movement (Margaria et al., 2016). Details of replication are not known except that CP interferes with the plant silencing defense only in the context of virus infection (Rossi et al., 2015).
The 82 kDa RdRP of BOLV is encoded by its single genome RNA segment (Donaire et al., 2016); a protein of similar size is encoded by members of the other genera. Mycoviruses can persist inside their fungal hosts without capsid or movement proteins, hence, it is assumed that members of genera Botoulivirus, Betabotoulivirus, Magoulivirus, Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus, Penoulivirus, Rhizoulivirus and Betarhizoulivirus may require only the RdRP for their replication, as has been shown for a member of the genus Penoulivirus (Wang et al., 2020).
Biology
Ourmia melon virus can easily be mechanically transmitted to a wide range of dicotyledonous plants (about 40 host species in 15 families have been reported), usually inducing systemic ringspots, mosaic and necrosis, with local lesions on some hosts. Tissue tropism is controlled by the KR-rich region at the amino terminus of the CP (Rossi et al., 2015). No vector has been identified but several species of weeds around infected fields are commonly infected, suggesting horizontal transmission of the virus. However, experimental transmission has not been obtained with several aphid species, the whiteflies Trialeurodes vaporariorum and Bemisia tabaci, or the mite Tetranychus urticae. Attempts at transmission through soil or irrigation water have been unsuccessful. Experimental seed transmission rates are 1–2% in Nicotiana benthamiana and N. megalosiphon. Members of different species in the genus Ourmiavirus occur in geographically diverse areas and in widely different hosts, though there are experimental hosts in common. Members of the other genera in the family infect fungi in different genera such as Botrytis (Donaire et al., 2016, Ruiz-Padilla et al., 2021, Wang et al., 2022), Sclerotinia (Marzano et al., 2016, Liang et al., 2020, Wang et al., 2020), Magnaporthe (Illana et al., 2017, Li et al., 2019, Liu et al., 2020, Zhou et al., 2021), Rhizoctonia (Picarelli et al., 2019), Epicoccum (Nerva et al., 2019b), Acremonium (Nerva et al., 2019b), Phaeoacremonium (Nerva et al., 2019b), Neofusicoccum (Nerva et al., 2019b), Cladosporium (Nerva et al., 2019b), Entoleuca (Velasco et al., 2019), Colletotrichum (Guo et al., 2019), Penicillium (Nerva et al., 2019a), Pyricularia (Ohkita et al., 2019), Phoma (Zhou et al., 2020), Aspergillus (Gilbert et al., 2019), Phomopsis (Hrabáková et al., 2017), Macrophomina (Wang et al., 2021) and Oidiodendron (Sutela et al., 2020), or have been associated with grapevine infected leaves infected with the oomycete Plasmopara viticola (Chiapello et al., 2020) or with the ascomycete Erysiphe necator (Galán-Cubero et al., 2022), soybean leaves (Marzano and Domier 2016), Pestalotiopsis (Chen et al., 2021), Fusarium (Zhao et al., 2020), Armillaria (Linnakoski et al., 2021), Heterobasidion (Sutela et al., 2021), Botryosphaeria (Lian et al., 2021) or apple plant tissue showing symptoms of apple decline (Wright et al., 2020).
Sclerotinia sclerotiorum ourmia-like virus 4 (SsOLV4) has been found in cytoplasm and mitochondria, and the infectious clone of this mycovirus could transfer among S. sclerotiorum strains via hyphal anastomosis, suggesting that only the RdRP-coding RNA segment is sufficient for transmission (Wang et al., 2020).
Antigenicity
Virions of Ourmia melon virus (i.e. the assembled CP) are good immunogens, as are the tubular structures associated with the MP. Antisera to these proteins do not react in Western blots to proteins in extracts of plants infected with members of either of the other two virus species in the genus Ourmiavirus.
Derivation of names
Botoulivirus, Betabotoulivirus: from Botrytis and ourmia-like
Botourmiaviridae: from Botrytis and ourmia-like
Magoulivirus: from Magnaporthe and ourmia-like
Ourmiavirus: from Ourmia (Urmia, Orumieh), a city in north-western Iran where the Ourmia melon virus was first found
Penoulivirus: from Penicillium and ourmia-like
Rhizoulivirus, Betarhizoulivirus: from Rhizoctonia and ourmia-like
Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus: from Sclerotinia and ourmia-like
Genus demarcation criteria
Members of the genus Ourmiavirus are clearly separated from members of the other genera based on their host (plants rather than fungi), the number of genomic segments (three segments vs one segment) and the presence of a MP and a CP (only for Ourmiavirus). For the other genera that include members infecting fungi, and possibly other hosts (Botoulivirus, Betabotoulivirus, Magoulivirus, Scleroulivirus, Betascleroulivirus, Gammascleroulivirus, Deltascleroulivirus, Epsilonscleroulivirus, Penoulivirus, Rhizoulivirus, and Betarhizoulivirus), genus demarcation is based on differences in RdRP amino acid sequence. Members of different genera in the family Botourmiaviridae share less than 70 % identity in their complete RdRP amino acid sequences.
Relationships within the family
There are twelve genera in the family Botourmiaviridae. Members of the family form a monophyletic, bootstrap supported group upon analysis of the RdRP gene, as do members of each of the twelve genera (Figure 4 Botourmiaviridae).
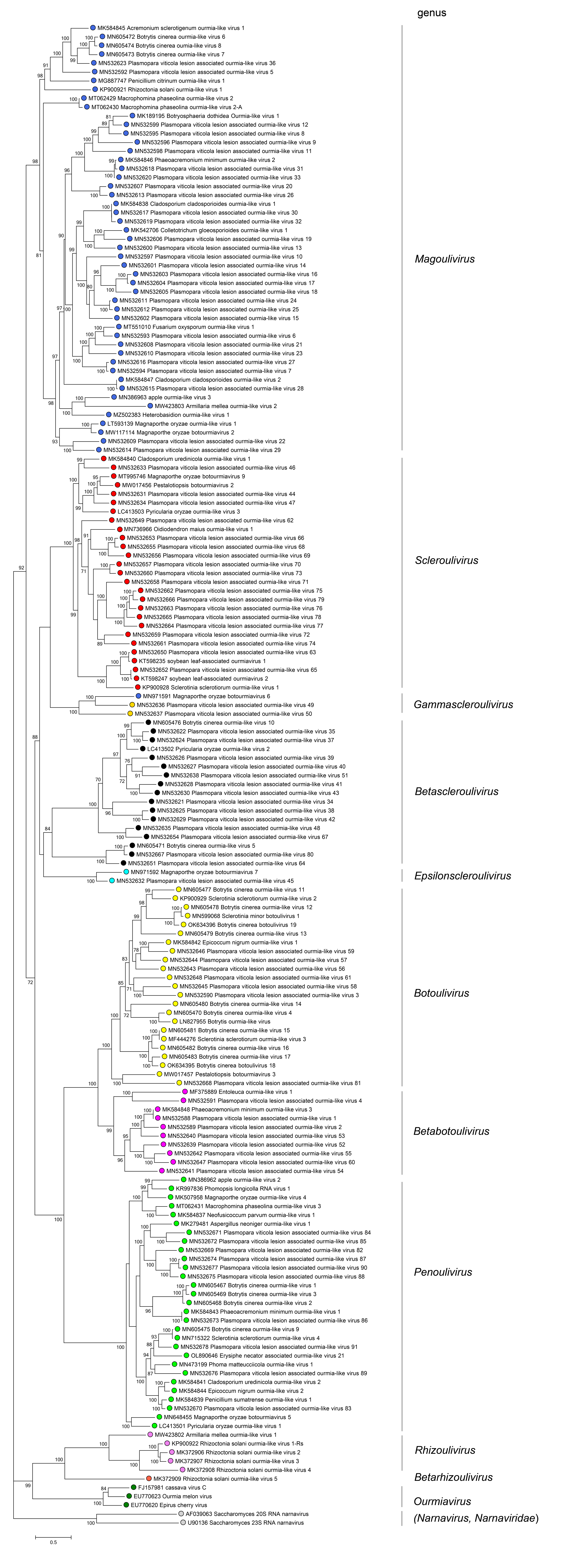 |
| Figure 4 Botourmiaviridae. Phylogenetic tree of members of the family Botourmiaviridae. A Maximum Likelihood phylogenetic tree was constructed based on the multiple amino acid sequence alignment (Clustal Omega) of the RNA-directed RNA polymerase (RdRP) using IQ-TREE (version 1.6.11) (Nguyen et al., 2015, Trifinopoulos et al., 2016) with the best-fit model “VT+F+R8” and 1,000 replicates ultrafast bootstrap (Minh et al., 2013). Viruses classified in the genus Narnavirus were used as outgroups. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Viruses of the genus Ourmiavirus are exceptional, having particles of unique morphology and a unique combination of phylogenetic affinities for the three different genomic RNAs. The MP encoded by RNA2 has clear similarities with the MPs of viruses in the family Tombusviridae (Rastgou et al., 2009). The CP shows distant affinities with the CPs of sobemoviruses, tombusviruses and luteoviruses (plant viruses) and nodaviruses (animal viruses) (Rastgou et al., 2009). The RdRP encoded by RNA1 has affinity with the RdRP of ourmia-like viruses from invertebrate hosts (Shi et al., 2016).

