Family: Hypoviridae
Sotaro Chiba, Leonardo Velasco, María A. Ayllón, Nobuhiro Suzuki, Shin-Yi L. Marzano, Lying Sun, Sead Sabanadzovic and Massimo Turina
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Massimo Turina ([email protected]))
Edited by: Sead Sabanadzovic
Posted: May 2018, updated March 2023
PDF: ICTV_Hypoviridae.pdf (2018 version)
Summary
Hypoviridae, comprising eight genera (Alphahypovirus, Betahypovirus, Gammahypovirus, Deltahypovirus, Epsilonhypovirus, Zetahypovirus, Thetahypovirus, Etahypovirus), is a family of capsidless viruses with positive-sense RNA genomes of 7.3–18.3 kb that possess either a single large open reading frame (ORF) or two ORFs. The ORFs appear to be translated from genomic RNA by non-canonical mechanisms, i.e., internal ribosome entry site-mediated and stop/restart translation. Hypovirids have been detected in ascomycetous and basidiomycetous filamentous fungi, and are considered to replicate in host Golgi body-derived, lipid vesicles that contain their dsRNA as the replicative form. Some hypoviruses induce hypovirulence to host fungi, while others do not.
Table 1. Hypoviridae. Characteristics of members of the family Hypoviridae.
| Characteristic | Description |
| Example | Cryphonectria hypovirus 1 (M57938), species Alphahypovirus cryphonectriae, genus Alphahypovirus |
| Virion | Capsidless (no true virions) |
| Genome | 7.3–18.3 kb of linear, positive-sense, unsegmented RNA |
| Replication | Replication and transcription occur cytoplasmically and for Cryphonectria hypovirus 1 in Golgi body-derived membranous vesicles |
| Translation | Directly from bi- or mono-cistronic genomic RNA containing a possible internal ribosomal entry site at the 5′-non-coding region. Cryphonectria hypovirus 1 ORF B is translated through re-initiation after ORF A stop codon |
| Host range | Fungi; those identified in arthropods need experimental confirmation as to host |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Duploviricetes, order Durnavirales: the family Hypoviridae includes eight genera (Alphahypovirus, Betahypovirus, Gammahypovirus, Deltahypovirus, Epsilonhypovirus, Zetahypovirus, Thetahypovirus, Etahypovirus) and 39 species |
Virion
Morphology
No true virions are associated with hypoviruses. In the case of Cryphonectria hypovirus 1 (CHV1, genus Alphahypovirus), pleomorphic vesicles 50–80 nm in diameter (Newhouse et al., 1983), devoid of any detectable viral structural proteins but containing replicative form dsRNA and polymerase activity (Fahima et al., 1994), are the only virus-associated structures that can be isolated from infected fungal tissue.
Physicochemical and physical properties
The Mr of vesicles is unknown. They have a buoyant density in CsCl of approximately 1.27–1.30 g cm−3 and sediment through sucrose as a broad component of approximately 200S (Dodds 1980). Their pH stability is unknown. The vesicles can be purified in pH 5.0 buffer and resuspended in pH 7.0 buffer. The pH optimum for polymerase activity in vitro is 8.0; the optimum Mg2+ concentration for polymerase activity is 5 mM. Activity decreases dramatically at pH less than 7.0 or more than 9.0 (Fahima et al., 1993). The vesicles are unstable when heated, or dispersed in lipid solvents. The optimal temperature for polymerase activity is 30 °C; temperatures over 40 °C abolishes polymerase activity. Deoxycholate at concentrations of more than 0.5% also abolishes polymerase activity (Fahima et al., 1993).
Nucleic acid
Hypovirus genomes are positive-sense RNA varying in size between 7.3 and 18.3 kb. The genome of Cryphonectria hypovirus 1 isolate EP713 (CHV1-EP713), the first completely sequenced hypovirus, is 12 712 nt. The coding (positive-sense) strand contains a short 3′-poly(A) tail, of 20–30 nt when analyzed as a component of the dsRNA (Shapira et al., 1991b).
In the absence of an identifiable virion RNA, hypoviruses were originally classified, along with several other fungal viruses, as dsRNA viruses. However, the RNA-directed RNA polymerase (RdRP) and RNA helicase of hypoviruses show sequence similarity and phylogenetic affinity to plant potyviruses, members of the expanded picorna-like supergroup with positive-sense RNA genomes (Koonin et al., 1991). In addition, the ability of positive-sense RNA, but not dsRNA, to initiate infection is not consistent with their current classification that groups them in order Durnavirales together with bona fide dsRNA viruses, such as the members of the family Partitiviridae. In fact, their phylogenetic position is not well supported statistically (Wolf et al., 2018). The predominant dsRNA form found in infected mycelia is regarded as a replicative intermediate or replicative form RNA. The accumulation of positive-sense RNA, by contrast, is low due to the RNA silencing antiviral defense response of the fungal host, but greatly increases in fungal strains in which the corresponding Dicer (dcl2) or Argonaute gene (agl2) has been disrupted (Segers et al., 2007, Sun et al., 2009). Small interfering RNAs (siRNAs) spanning the entire genomic RNA are produced in a non-random fashion (Zhang et al., 2008). The presence of shorter-than-full-length, internally deleted, defective interfering (DI) and defective (D) replicative form dsRNA molecules is common among some members, and replicative forms of satellite-like RNAs are present in other members (Shapira et al., 1991a, Yuan and Hillman 2001). The host RNA silencing pathway has been reported to promote DI RNA production (Nuss 2011). No function has been ascribed to any ancillary dsRNA. The 5′-terminus of the positive-sense strand of dsRNA from Cryphonectria hypovirus 1-EP713 (CHV1-EP713) is blocked (Hiremath et al., 1986) but the nature of the blocking group is unknown. The 5′-terminus of the negative-sense strand is unblocked. Both 5′-termini of dsRNA from Cryphonectria hypovirus 3-GH2 (CHV3-GH2) are unblocked (Tartaglia et al., 1986).
Proteins
No hypovirus structural proteins have been described.
Lipids
Host-derived lipids make up the vesicles that encapsulate the viral dsRNA (Newhouse et al., 1990).
Carbohydrates
Carbohydrates similar to those involved in fungal cell wall synthesis are associated with vesicles (Hansen et al., 1985).
Genome organization and replication
A 5′-leader of 300–500 nt, including several AUG triplets, precedes the AUG codon that initiates the first long ORF (Figure 1.Hypoviridae). Translational initiation for the first ORF on the genomic RNA is mediated by an internal ribosome entry site (IRES) residing in the 5′-non-coding region (NCR) and extending to the coding domain in the case of CHV1 (Chiba et al., 2018), although 5′-NCR sequences and predicted secondary structures are highly variable between members of hypovirus species (Mu et al., 2011). The viral coding region may consist of a single long ORF, or may be divided into two ORFs. If two ORFs are present (e.g., CHV1 and CHV2), the shorter, 5′-proximal ORF is designated ORF A. Its product may or may not be autocatalytically cleaved, depending on the virus. The UAA termination sequence at the end of ORF A is part of the pentanucleotide UAAUG present in all members with two ORFs investigated to date. In the case of hypoviruses with a bicistronic genome organization, the stop/restart translation mechanism is involved for the translation of their downstream ORFs. The pentanucleotide, UAAUG for CHV1 and CHV2 at the junction of the two ORFs (Figure 1.Hypoviridae), plays a critical role (Guo et al., 2009). The N-terminal product of ORF B contains a papain-like cysteine protease domain that is autocatalytically released from the growing polypeptide chain (e.g., p48 for CHV1-EP713 or p52 for CHV2-NB58) (Shapira and Nuss 1991, Hillman et al., 1994). No further processing in vitro has been experimentally demonstrated for the remaining 314 kDa polypeptide (2747 amino acids) from this ORF, although processed products have been detected in vivo (Fahima et al., 1994).
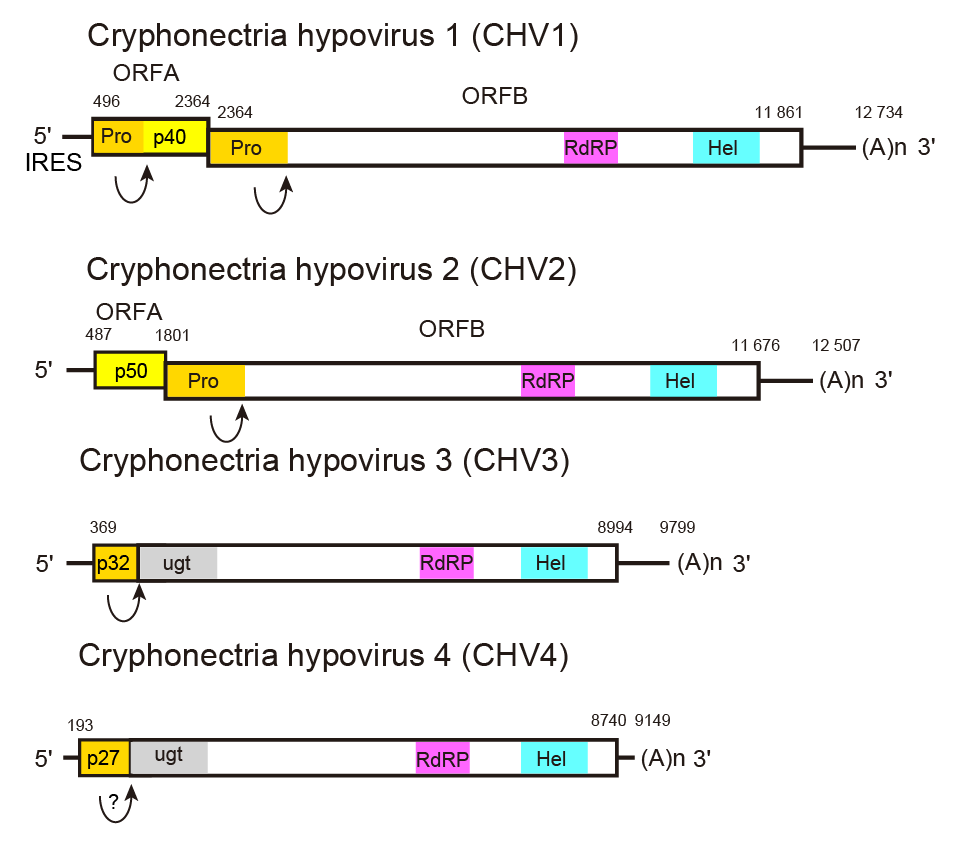 |
| Figure 1.Hypoviridae. Genome organization of selected members of the family Hypoviridae. Arrows represent known or suspected sites of autoproteolysis. Cryphonectria hypovirus 4 has a genome organization similar to that of Cryphonectria hypovirus 3, but it is unknown whether the former undergoes autoproteolysis. The abbreviations Pro, RdRP, Hel, and ugt refer to the protease, RNA-directed RNA polymerase, RNA helicase, and UDP-glucose/sterol glucosyltransferase domains, respectively. |
Among the hypoviruses infecting Botrytis cinerea, a papain-like protease domain, involved in hypovirus polyprotein processing, is also coded by Botrytis cinerea hypovirius 3 and Botrytis cinerea hypovirus 5 genomes; whereas a 2-A like protease domain (DIEQNPGP, aa 1076 to 1083) is encoded by Botrytis cinerea hypovirus 2 (Ruiz-Padilla et al., 2021).
Phylogenetic relatedness to members of the positive-sense RNA family Potyviridae has been demonstrated by comparisons of protease, polymerase and helicase domains, although these domains are positioned differently in the polyproteins encoded by genomes of viruses belonging to these two families (Koonin et al., 1991). The 5′-proximal portions of hypovirus genomes are prone to recombination and show greater molecular variability than their 3′-proximal part (Wang et al., 2013, Marzano et al., 2015, Du et al., 2017).
Synthesis of both positive- and negative-sense viral RNA is believed to occur cytoplasmically in host-derived lipid vesicles that contain linear dsRNA, regarded as the replicative form of hypoviral positive-sense RNA. The polymerase associated with vesicles transcribes ssRNA molecules in vitro that correspond in size to full-length dsRNA. Approximately 80% of the polymerase products in vitro are of positive-sense polarity (Fahima et al., 1993). Except for CHV2 p50, hypoviral proteins are synthesized as part of a polyprotein. The polyprotein is autocatalytically cleaved by viral proteases such as CHV1 p29 and p48, and CHV2 p52 (Figure 2.Hypoviridae). CHV1 p29 enhances the replication in cis and in trans by suppressing antiviral RNA silencing (Segers et al., 2007), regardless of whether expressed from the viral genome or transgenically. The other CHV1 ORF A-coded p40, a basic protein, also enhances the replication in cis only (Suzuki and Nuss 2002). However, the ORF A coding domain, except the first 22 codons, is dispensable for CHV1 viability (Suzuki et al., 2000). The autocatalytic processing of the p29 and p48 contributes to virus RNA accumulation but are not essential for virus replication. (Jensen and Nuss 2014).
 |
| Figure 2. Hypoviridae. Genome organization of members of the family Hypoviridae. One representative from each genus is shown. Black lines represent the genome, whereas boxes represent the open reading frames (ORF) with the main conserved domains indicated. Protein products and processing are indicated for Cryphonectria hypovirus 1. The abbreviations Pro, RdRP, Hel and UGT refer to the protease, RNA-directed RNA polymerase, RNA helicase and UDP-glucose/sterol glucosyltransferase domains, respectively. IRES= internal ribosomal entry site. |
Proteins
No hypovirus structural proteins have been described. Functions have been assigned to several non-structural polypeptides encoded by hypoviruses. The 5′-proximal coding domain, ORF A, of CHV1-EP713 RNA encodes a papain-like protease, p29, and a highly basic protein, p40, derived, respectively, from the N-terminus and C-terminus of polyprotein p69, by a p29-mediated cleavage event (Choi et al., 1991a, Choi et al., 1991b) (Figure 1.Hypoviridae). A presumptive non-structural protein identified in vitro and in vivo, p29, has been shown by DNA-mediated transformation to contribute to suppression of host pigmentation, reduced sporulation, reduced laccase accumulation (Craven et al., 1993), to enhance vertical virus transmission (Suzuki et al., 2003), to serve as a suppressor of host RNA silencing (Zhang and Nuss 2008), to be an integral membrane protein targeting membranes(Jacob-Wilk et al., 2006) and to promote RNA recombination of a co-infecting mycoreovirus (dsRNA virus) (Sun and Suzuki 2008). Protein p40 enhances viral RNA accumulation (Suzuki and Nuss 2002). RNA polymerase activity is associated with isolated vesicles from a CHV1-EP713 infected fungal isolate (Fahima et al., 1993, Fahima et al., 1994). The calculated mass of the CHV1 ORF B product, which contains a second papain-like protease, p48, along with putative RNA polymerase and helicase domains, is approximately 362 kDa (3165 amino acids) based on its deduced aa sequence, but no protein of that mass has yet been detected from vesicles. Smaller virus-encoded proteins (about 87 kDa) have been identified in the vesicle-associated polymerase complex, suggesting extensive processing of replication-associated proteins (Fahima et al., 1994). However, exact cleavage sites for these proteins remain unknown. Other, as-yet-unidentified, viral and/or host proteases may contribute to the polyprotein processing. Note that 2A-like self-processing peptide motifs are detectable in some hypoviruses, although whether they are functional in infected fungal cells is still unclear (Petrzik et al., 2016, Hisano et al., 2017). CHV3 and Cryphonectria hypovirus 4 (CHV4) each encode a putative UDP glycosyltransferase-encoding domain that is not present in CHV1 or Cryphonectria hypovirus 2 (CHV2), although the specific function of this domain has not been examined (Smart et al., 1999, Linder-Basso et al., 2005).
Biology
Hypovirues were originally identified in the chestnut blight fungus, Cryphonectria parasitica, isolated from chestnut-growing areas of Europe, North America and Asia (Peever et al., 1998, Bryner et al., 2012). DsRNA-containing vesicles have been associated with abnormal Golgi apparatus in freeze-substituted thin sections (Newhouse et al., 1990). No nuclear or mitochondrial associations, nor virus-associated inclusions, have been noted. Members that infect the chestnut blight fungus, Cryphonectria parasitica, induce reduced host virulence (hypovirulence) on chestnut trees and altered fungal morphology in culture, but many other family members have little or no discernible effect on the fungal host. Other fungi infected by hypoviruses are Sclerotinia sclerotiorum (Xie et al., 2011, Hu et al., 2014, Khalifa and Pearson 2014, Marzano et al., 2015), Valsa ceratosperma (Yaegashi et al., 2012), Phomopsis longicolla (Koloniuk et al., 2014), Fusarium spp. (Wang et al., 2013, Li et al., 2015, Osaki et al., 2016, Li et al., 2017), Macrophomina phaseolina (Marzano et al., 2016), and Agaricus bisporus (Deakin et al., 2017). Some hypoviruses have been detected in RNA extracts from invertebrates, but their specific host is not known. Infection of fungal mycelium is known only to occur through fusion, or anastomosis of infected with uninfected hyphae. The transmission rate through asexual spores (conidia) varies from a few percentages to close to 100%. Transmission through sexual spores (ascospores) is not known to occur. Transmission via cell-free extracts has not been demonstrated but transfection of protoplasts with full-length synthetic transcripts derived from cDNA clones has been successful for hypoviruses CHV1-EP713, CHV1-Euro7, CHV1-EP721, and Sclerotinia sclerotiorum hypovirus 2-Lactuca (SsHV2-L), a member of the genus Thetahypovirus (Chen et al., 1994, Chen and Nuss 1999, Lin et al., 2007, Marzano et al., 2015).
CHV1-EP713 can be transmitted by protoplast fusion to Valsa mali, the causal agent of apple valsa canker disease, conferring hypovirulence to the host (Yang et al., 2021).
Antigenicity
No antibody has ever been raised from virus-associated vescicles preparations. Antibodies directed against CHV1-EP713-encoded p29 have been used to demonstrate its association with the trans-Golgi network membranes (Jacob-Wilk et al., 2006). Antibodies directed against the conserved RdRP domain of ORF B, expressed in bacteria, have been used to identify an 87 kDa protein in a CHV1-EP713 infected fungal strain (Fahima et al., 1994).
Derivation of names
Hypoviridae: from hypovirulence.
Genus demarcation criteria
Genera are distinguished based on phylogenetic trees derived from alignments along the whole sequence of the polyproteins that include the RdRP domain. Members of different genera belong to monophyletic clades present in the tree depicted in Figure 3.Hypoviridae.
Relationships within the family
Eight monophyletic groups can be identified by phylogenetic analysis of complete polyprotein sequences corresponding to the eight genera in the family (Figure 3.Hypoviridae). Among the 44 RdRP sequences used to build the tree, pairwise analysis showed that five viruses share more than 80% identity to another virus and therefore belong to the same species.
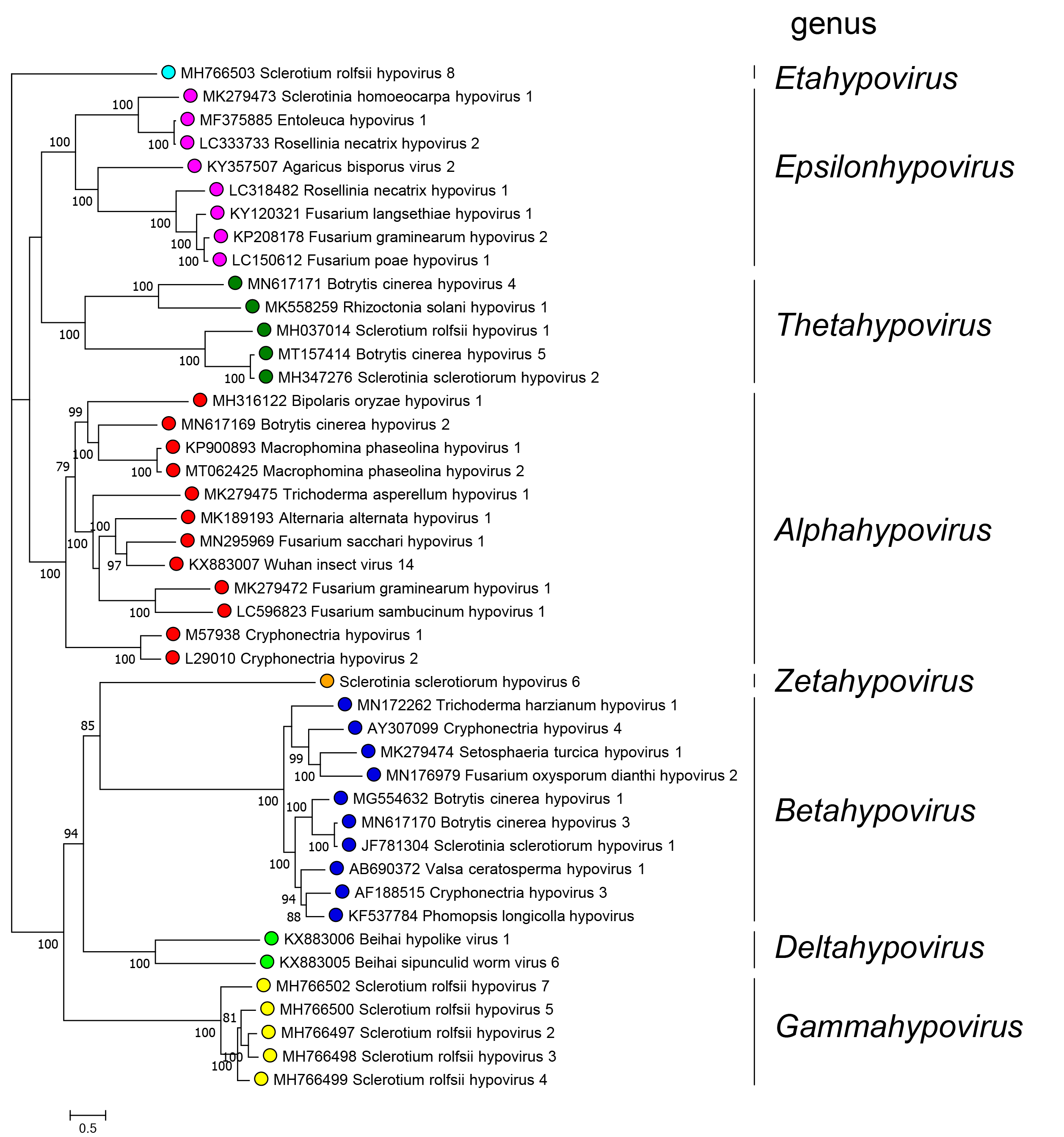 |
| Figure 3.Hypoviridae. . Phylogenetic relationships among members of the family were derived by performing a maximum-likelihood analysis implemented with the IQ-TREE software (Nguyen et al., 2015) using the Model Finder to calculate the best substitution model (Kalyaanamoorthy et al., 2017) to derive a tree based on alignments of the entire polyprotein containing the RdRP domain of 44 viruses with complete or almost complete coding sequences. Proteins were aligned with MAFFT version 7 (Katoh et al., 2017). A maximum likelihood phylogenetic tree was generated using IQ-TREE with the best-fit model LG+F+I+G4. Numbers at the nodes indicate bootstrap support (1000 replicates) where this was >70%. Coloured dots at branch tips indicated the genus to which a virus belongs. |
Relationships with other taxa
The possession of an undivided positive-sense RNA genome, expressed via long polyprotein precursors, demonstrates an affinity with viruses of the “picornavirus supergroup” (Koonin et al., 1991). Deduced aa sequences of polymerase, helicase and protease motifs of members of the family Hypoviridae suggest that their closest relatives are the members of the family Fusariviridae (Kwon et al., 2007, Zhang et al., 2014) and more distantly to members of the genus Bymovirus in the family Potyviridae (Koonin et al., 1991). However, fusariviruses have a different genome organization and relative position of the polymerase and helicase domains. Furthermore, several groups of viruses that putatively code for a coat protein have recently been detected in metatranscriptomic studies that are basal to members of the family Hypoviridae in phylogenetic analyses, suggesting that ancestors of hypoviruses may have encoded a CP (Neri et al., 2022).

