Family: Phasmaviridae
Jens H. Kuhn and Holly R. Hughes
The citation for this ICTV Report chapter is the summary published as Kuhn et al. (2024):
ICTV Virus Taxonomy Profile: Phasmaviridae 2024, Journal of General Virology (2024) 105: 002002
Corresponding author: Holly R. Hughes ([email protected])
Edited by: Holly R. Hughes and Evelien Adriaenssens
Posted: May 2024
Summary
Phasmaviridae is a family for negative-sense RNA viruses with genomes of about 9.7–15.8 kb (Table 1 Phasmaviridae). The family includes seven genera (Cicadellivirus, Feravirus, Hymovirus, Jonvirus, Orthophasmavirus, Sawastrivirus, and Wuhivirus). These viruses are maintained in and/or transmitted by blattodean, coleopteran, dipteran, hemipteran, hymenopteran, neuropteran, and odonatan insects. Phasmavirids produce enveloped virions containing three single-stranded RNA segments with open reading frames (ORFs) that encode a nucleoprotein (N), a glycoprotein precursor (GPC), and a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain.
Table 1 Phasmaviridae. Characteristics of members of the family Phasmaviridae.
| Characteristic | Description |
| Example | Kigluaik phantom virus (small [S] segment: KJ434184; medium [M] segment: KJ434183; large [L] segment: KJ434182), species Orthophasmavirus kigluaikense |
| Virion | Enveloped, spherical or pleomorphic (60–120 nm in diameter), or tubular (60 × 600 nm) virions with heterodimeric surface spikes |
| Genome | Three negative-sense RNA molecules (segments): S: 1.3–2.9 kb; M: 2.0–6.6 kb; and L: 6.3–7.7 kb |
| Replication | Ribonucleoprotein (RNP) complexes contain anti-genomic RNA and serve as coding templates for the synthesis of genomic RNA |
| Translation | From non-polyadenylated mRNAs with a 5′-cap structure derived by cap-snatching from cellular mRNAs |
| Host range | Insects |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Elliovirales: the family includes 7 genera and 30 species |
Viruses assigned to each of the seven genera form a monophyletic clade based on phylogenetic analysis of L protein/RdRP sequences. Viruses from all seven genera share one or more of the following characteristics: (i) enveloped spherical, pleomorphic, or tubular virions; (ii) tri-segmented negative-sense RNA genome; (iii) genomic sequence complementarity at the 5′- and 3′-ends; and (iv) capped but not polyadenylated virus mRNAs. All genera have insect hosts (Table 2 Phasmaviridae).
Table 2. Phasmaviridae. Insect hosts of phasmavirid genera.
| Genus | Host order(s) | Reference |
| Cicadellivirus | Hemiptera | (Ottati et al., 2020) |
| Feravirus | Diptera, Hemiptera and Neuroptera | (Marklewitz et al., 2015, Käfer et al., 2019) |
| Hymovirus | Hymenoptera | (Käfer et al., 2019) |
| Jonvirus | Diptera | (Marklewitz et al., 2015) |
| Orthophasmavirus | Blattodea, Coleoptera, Diptera, Hemiptera, Hymenoptera and Odonata | (Ballinger et al., 2014, Li et al., 2015, Schoonvaere et al., 2016, Shi et al., 2016, Makhsous et al., 2017, Shi et al., 2017, Käfer et al., 2019, Öhlund et al., 2019, Batson et al., 2021, Truong Nguyen et al., 2022, Zhang et al., 2022, Matsumura et al., 2024) |
| Sawastrivirus | Hemiptera | (Li et al., 2015) |
| Wuhivirus | Hemiptera | (Li et al., 2015) |
Virion
Morphology
Only known for members of the genera Feravirus and Jonvirus (see Feravirus and Jonvirus genus pages).
Physicochemical and physical properties
Only known for members of the genera Feravirus and Jonvirus (see Feravirus and Jonvirus genus pages).
Nucleic acid
Phasmavirids have tri-segmented negative-sense RNA genomes (small [S], medium [M], and large [L] segments) (Table 3 Phasmaviridae). Viral mRNAs are not polyadenylated and contain a 5′-methylated cap and several non-templated nucleotides at the 5′-end that are derived from host cell mRNAs via cap-snatching.
Table 3 Phasmaviridae. Viral RNA segments of typical genus members
| Genus | Virus | RNA segment (bases) | ||
| L | M | S | ||
| Cicadellivirus | Scaphoideus titanus bunya-like virus 1 | 7740 | 4361 | 1458 |
| Feravirus | Ferak virus | 6936 | 4274 | 1527 |
| Hymovirus | hymenopteran phasma-related virus OKIAV250 | 6615 | 4199 | 1504 |
| Jonivirus | jonchet virus | 6904 | 5449 | 1745 |
| Orthophasmavirus | Kigluaik phantom virus | 6697 | 2792 | 2212 |
| Sawastrivirus | Sānxiá water strider virus 2 | 7151 | 4213 | 1628 |
| Wuhivirus | Wǔhàn insect virus 2 | 7243 | 6515 | 1700 |
Proteins
Phasmavirids typically express three structural proteins. The most abundant structural protein in a phasmavirion is N, which encapsidates the phasmavirid genomic segments. The least abundant is the L protein, which mediates viral genome replication and transcription. Two glycoprotein subunits, GN and GC, are produced from GPC and mediate cell entry of phasmavirions (Marklewitz et al., 2015).
Lipids
Likely derived from the Golgi apparatus and/or the host cell plasma membrane and therefore likely composed of phospholipids, glycolipids, fatty acids, and sterols.
Carbohydrates
Only known for members of the genera Feravirus and Jonvirus (see Feravirus and Jonvirus genus pages).
Genome organization and replication
The S RNA encodes N and in some viruses a nonstructural protein (NSs) and/or another protein of unknown function. The M RNA encodes GPC and in some viruses a nonstructural protein (NSm). The L RNA encodes L protein with its RdRP, helicase, and endonuclease domains (Marklewitz et al., 2015, Käfer et al., 2019) (Figure 1 Phasmaviridae).
 |
| Figure 1 Phasmaviridae. Genome organization of representative phasmavirids. The 5′- and 3′- ends of all segments are complementary at their termini, likely promoting the formation of panhandle RNP complexes within the virion. GPC, glycoprotein precursor gene; L, large (protein) gene; N, nucleoprotein gene. |
Biology
Classified phasmavirids are maintained in and/or transmitted by blattodean, coleopteran, dipteran, hemipteran, hymenopteran, neuropteran, and odonatan insects in Africa, Asia, Europe, Northern America, and South America (da Silva Ferreira et al., 2020, Ottati et al., 2020, da Silva Neves et al., 2021, He et al., 2021, Hermanns et al., 2023, Starchevskaya et al., 2023, Yang et al., 2023, Ballinger et al., 2014, Li et al., 2015, Marklewitz et al., 2015, Schoonvaere et al., 2016, Shi et al., 2016, Makhsous et al., 2017, Shi et al., 2017, Käfer et al., 2019, Öhlund et al., 2019, Batson et al., 2021, Hermanns et al., 2023). Putative phasmavirids have been associated with these insects and branchiopods, collembolan hexapods, myriapods, zorapteran insects, as well as in eulipotyphla, mammals, plants, and fungi in Asia, Europe, Northern America, and Oceania (Li et al., 2015, Käfer et al., 2019, Kobayashi et al., 2020, Lay et al., 2020, Dou et al., 2021, Chen et al., 2022, French et al., 2023, Dong et al., 2024, Medd et al., 2018, Wallace et al., 2021, Tian et al., 2022, Truong Nguyen et al., 2022, Zhang et al., 2022, Litov et al., 2023, Matsumura et al., 2024).
Derivation of names
aedis: after host genus Aedes
anophelae: after host genus Anopheles
bastukasense: after Lithuanian Barstukas, a mythical dragon-like creature; the suffix “ense” is incorrectly used and will be corrected
chrysis: after host genus Chrysis
chrysurae: after host genus Chrysura
Cicadellivirus: after host family Cicadellidae
coleopterus: after host order Coleoptera
coredoense: after Coredo, Italy
culicis: after host genus Culex
eboris: after the Latin ebur, meaning ivory, a reference to Côte d'Ivoire, where jonchet virus was discovered
ferakinum: after Ferak, the name of a female cartoon character belonging to a botanically-bred race of plant people
Feravirus: after Ferak virus
flenense: after Flen, Sweden
fushunense: after Fǔshùn (抚顺市), China
gandaense: after Ganda, the Latin name for Ghent, Belgium
guaguaense: after Guagua, Colombia
hemipterus: after host order Hemiptera
hubeiense: after Húběi Province (湖北省), China
Hymovirus: after host order Hymenoptera
insecti: after the Latin insectum, meaning insect
Jonvirus: after jonchet virus
kigluaikense: after the Kigluaik mountains, USA
miglotasense: after Lithuanian miglotas, meaning obscure, foggy, misty. The suffix “ense” is incorrectly used and will be corrected
neuropterus: after the host order Neuroptera
niuklukense: after Niukluk River, USA
odonatus: after host order Odonata
Orthophasmavirus: from the Ancient Greek ὀρθός (orthós), meaning straight, right, proper, and phantom midges, host of the first described viruses of the family
Phasmaviridae: after phantom midges, hosts of the first described viruses of the family
philoctetis: after host genus Philoctetes
sanxiaense: after Sānxiá (三峡), China
Sawastrivirus: after Sānxiá water strider virus
scaphoidei: after host genus Scaphoideus
sogatellae: after host genus Sogatella
wuchangense: after Wǔchāng (武昌), China
wuhanense: after Wǔhàn (武汉), China
Wuhivirus: after Wǔhàn insect virus 2
Genus and species demarcation criteria
The availability of at least coding-complete sequences of all three genome segments may be sufficient for phasmavirid classification in the absence of a cultured isolate. Viruses within the same genus form a monophyletic clade separate from those of other phasmavirid genera. The phasmavirid species demarcation criterion is <95% identity in the amino acid sequence of the L protein.
Relationships within the family
Phylogenetic relationships across the family have been estimated using phylogenetic trees generated from complete L amino acid sequences. The family is divided into two major clades: one including the genera Cicadellivirus and Orthoplasmavirus, and the other including the genera Feravirus, Hymovirus, Jonvirus, Sawastrivirus, and Wuhivirus (Figure 2 Phasmaviridae).
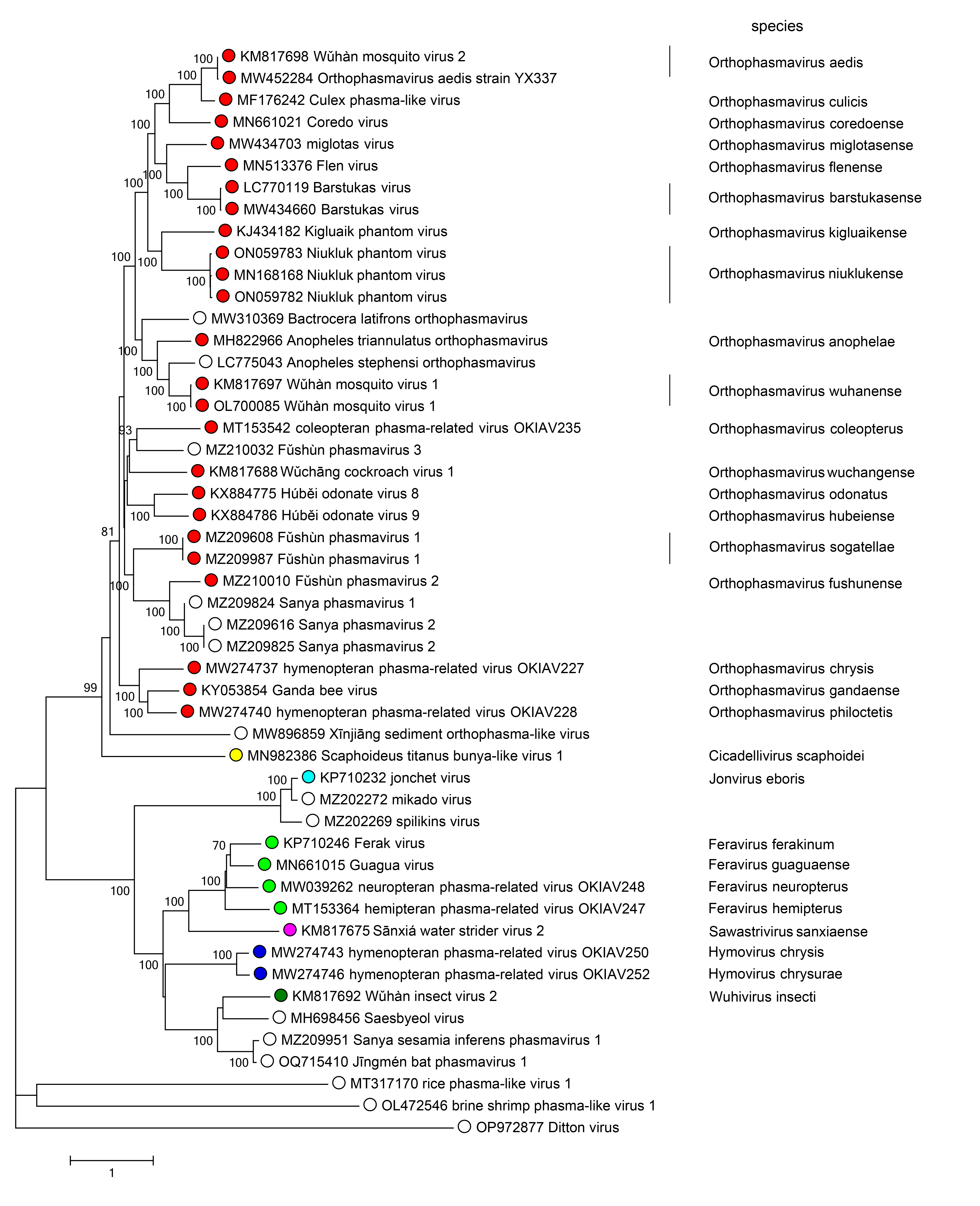 |
| Figure 2 Phasmaviridae. Phylogenetic analysis of phasmavirid L protein sequences. Publicly available coding-complete phasmavirid genomes were downloaded from NCBI GenBank. Coding sequences of the L segment were codon-aligned using Clustal W (Mega v10). The resulting amino acid sequences were used for phylogenetic inference by producing a neighbor-joining tree with a Poisson correction and a gamma distribution of variation between sites in MEGA7. Branches with <70% bootstrap support are not labeled. |
Relationships with other taxa
Phasmavirids are closely related to ellioviral crulivirids, fimovirids, hantavirids, peribunyavirids, tulasvirids, and tospovirids (Wolf et al., 2018, Herath et al., 2020, Olendraite et al., 2023).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| aphid bunyavirus 1 | L: OL752429; M: OL752430; S: OL752431; | ABV1 | (An et al., 2022) |
| beetle phasma-like virus | M: OQ986862* | (French et al., 2023) | |
| blattodean phasma-related virus OKIAV238 | L: MT153528*; S: MT153470 | (Käfer et al., 2019) | |
| blattodean phasma-related virus OKIAV239 | L:MT153445; S: MT153501 | (Käfer et al., 2019) | |
| brine shrimp phasma-like virus 1 | L: OL472546* | (Dong et al., 2024) | |
| Carapha virus | L: LC552039; M: LC552040; S: LC552041* | (Kobayashi et al., 2020) | |
| coleopteran phasma-related virus OKIAV236 | L: MT153461; M: MT153477*; S: MT153460 | (Käfer et al., 2019) | |
| coleopteran phasma-related virus OKIAV241 | L: MT153394; M: MT153367 | (Käfer et al., 2019) | |
| coleopteran phasma-related virus OKIAV243 | L: MT153444; M: MT153543*; S: MT153540 | (Käfer et al., 2019) | |
| collembolan phasma-related virus OKIAV223 | L: OP972877 | (Käfer et al., 2019) | |
| dipteran phasma-related virus OKIAV224 | L: MT153491; M: MT153552 | (Käfer et al., 2019) | |
| dipteran phasma-related virus OKIAV225 | L: MT153492; M: MT153488 | (Käfer et al., 2019) | |
| Ditton virus | L: MF893264 | (Medd et al., 2018) | |
| Drosophila North Esk phasmavirus | L: OR6505709; M: OR605710; S: OR605711 | (Wallace et al., 2021) | |
| Hángzhōu Frankliniella intonsa phasmavirus 1 | L: MZ209660; S: MZ209661 | ||
| Hángzhōu phasmavirus 1 | L: MZ209649 | ||
| Hángzhōu zicrona caerulea phasmavirus 1 | L: MZ209709 | ||
| hemipteran phasma-related virus OKIAV245 | L: MW288188*; M: MW288236*; S: MW288240 | (Käfer et al., 2019) | |
| Hénán orthophasma-like virus 1 | L: MW896857 | (Chen et al., 2022) | |
| Hénán sediment orthophasma-like virus 2 | L: MW896858* | (Chen et al., 2022) | |
| hymenopteran phasma-related virus OKIAV229 | L: MT153401*; M: MT153520; S: MT153354 | (Käfer et al., 2019) | |
| hymenopteran phasma-related virus OKIAV230 | L: MT153451*; M: MT153432; S: MT153360* | (Käfer et al., 2019) | |
| hymenopteran phasma-related virus OKIAV231 | L: MT153416*; S: MT153381* | (Käfer et al., 2019) | |
| hymenopteran phasma-related virus OKIAV232 | L: MW288175*; M: MW288201; S: MW288193 | (Käfer et al., 2019) | |
| hymenopteran phasma-related virus OKIAV233 | L: MW288196*; M: MW288241; S: MW288223 | (Käfer et al., 2019) | |
| hymenopteran phasma-related virus OKIAV234 | L: MW288198*; M: MW288232*; S: MW288212 | (Käfer et al., 2019) | |
| hymenopteran phasma-related virus OKIAV244 | L: MT153462*; M: MT153517; S: MT153490 | (Käfer et al., 2019) | |
| Jīngmén bat phasmavirus 1 | L: OQ715410 | (Chen et al., 2023) | |
| lepidopteran phasma-related virus OKIAV246 | L: MT153437*; M: MT153453*; S: MT153525 | (Käfer et al., 2019) | |
| Medvezhye Chrysops phasma-like virus | L: OR724686*; M: OR724687; S: OR724688 | (Litov et al., 2023) | |
| Medvezhye Tabanus phasma-like virus | S: OR724697* | (Litov et al., 2023) | |
| millipede phasma-like virus | M: OQ986858* | (French et al., 2023) | |
| millipede phasma-like virus 2 | M: OQ986859* | (French et al., 2023) | |
| millipede phasma-like virus 3 | M: OQ986860* | (French et al., 2023) | |
| moss associated phasma-like virus | L: OQ987804* | (French et al., 2023) | |
| moth phasma-like virus | M: OQ986861* | (French et al., 2023) | |
| Mucor phasmavirus A | L: MK231068* | ||
| Nanning phasm tick virus 1 | L: ON746495 | ||
| Notori virus | L: MF893255 | (Medd et al., 2018) | |
| pangolin Phasmaviridae sp. B5 | M: OP474158 | (Tian et al., 2022) | |
| Pectinophora gossypiella virus 2 | M: MZ361082; S: MZ361083 | (Dou et al., 2021) | |
| punctatus phasmavirus | (Konstantinidis et al., 2022) | ||
| Razdolnyj Hybomitra phasma-like virus | S: OR724689 | (Litov et al., 2023) | |
| rice phasma-like virus 1 | L: MT317170; S: MT317171 | ||
| Saesbyeol virus | L: MH698456; M: MH823676; S: MH698457 | ||
| Sanya sesamia inferens phasmavirus 1 | L: MZ209951; M: MZ209952; S: MZ209953* | ||
| Shuāngào insect virus 2 | L: KM817680*; M: KM817715; S: KM817740 | (Li et al., 2015) | |
| Wǔfēng shrew phasmavirus 1 | L: OQ715411* | (Chen et al., 2022) | |
| Xīnjiāng sediment orthophasma-like virus | L: MW896859; M: MW896860; S: MW896861 | (Chen et al., 2022) | |
| zorapteran phasma-related virus OKIAV242 | L: MT153351*; M: MT153487* | (Käfer et al., 2019) |
*incomplete sequence
Virus names and virus abbreviations are not official ICTV designations.
Additional unclassified phasmavirids that are probable members of established genera are listed under individual genus descriptions.

