Family: Peribunyaviridae
Genus: Shangavirus
Distinguishing features
Shangavirus RNA has been found in a mixed-insect pool from China containing both chrysopids (green lacewings) and moth flies (Psychoda alternata) (Li et al., 2015). There currently is no cultured shangavirus isolate.
Virion
Morphology
Not described.
Nucleic acid
The shangavirus genome consists of three segments of negative-sense RNA (Li et al., 2015). Analysis of the terminal sequences has yet to be completed.
Proteins
In silico analysis of shangavirus putative ORF sequences suggests shangaviruses encode a protein with RNA-directed RNA polymerase and endonuclease functions (L), the glycoproteins Gn and Gc, the non-structural protein NSm, and the nucleoprotein N, but not the non-structural protein NSs (Table 2 Peribunyaviridae) (Li et al., 2015).
Lipids
Not described.
Carbohydrates
There are 1 and 9 N-glycosylation sites on Gn and Gc, respectively (Li et al., 2015).
Genome organization and replication
The shangavirus genome is similar to the genomes of other peribunyaviruses (Figure 1 Shangavirus). The S, M, and L segments putatively encode N, Gn and Gc, and L proteins, respectively (Li et al., 2015).
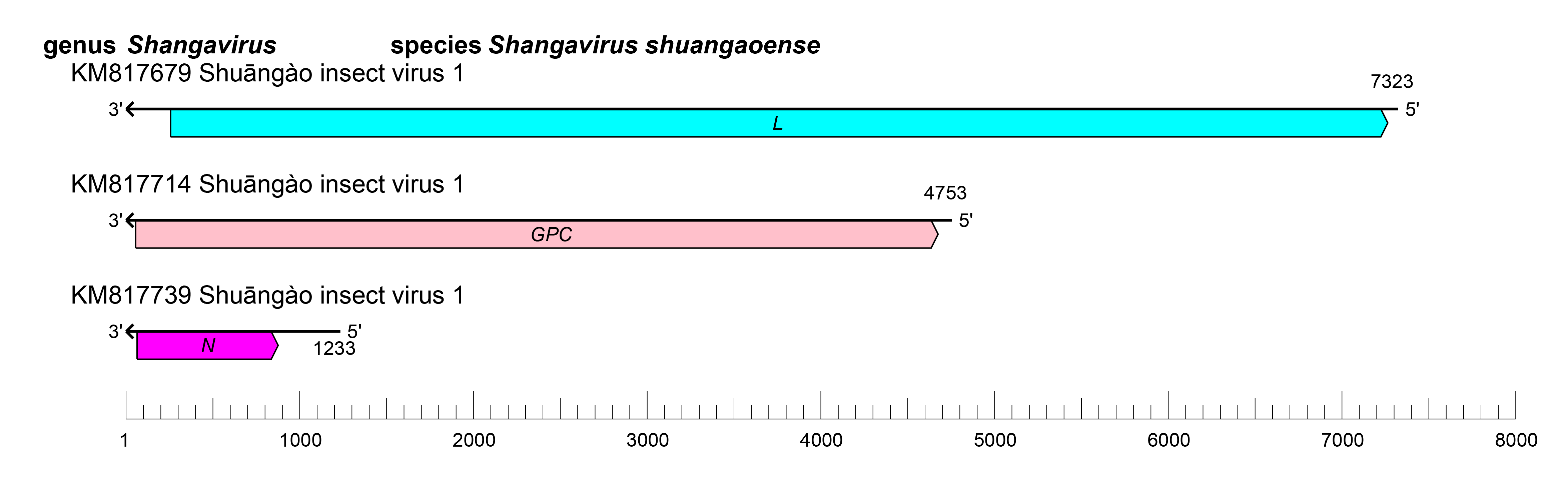 |
| Figure 1 Shangavirus. Shangavirus coding strategy. vcRNAs are depicted in 3′→5′ direction and mRNAs are depicted in a 5′→3′ direction. Coloured boxes on the mRNAs depict ORFs that encode the N, nucleocapsid protein; Gn and Gc, external glycoproteins; L, large protein, as well as the non-structural protein NSm. |
The shangavirus mode of replication is unknown.
Biology
Unknown, but suggested to be an insect-specific virus.
Species demarcation criteria
Not defined as the genus currently includes only a single species.

