Family: Peribunyaviridae
William M. de Souza, Charles H. Calisher, Jean Paul Carrera, Holly R. Hughes, Márcio R. T. Nunes, Brandy Russell, Natasha L. Tilston-Lunel, Marietjie Venter and Han Xia (夏菡)
The citation for this ICTV Report chapter is the summary published as de Souza et al., (2024) ICTV Virus Taxonomy Profile: Peribunyaviridae 2024 Journal of General Virology (2024) 105: 002034
Corresponding author: Holly R. Hughes ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2019, updated September 2021, October 2022, May 2023, September 2024
Summary
Peribunyaviruses are enveloped and possess three distinct, negative-sense RNA segments comprising 10.7−12.5 kb in total (Table 1 Peribunyaviridae). The family Peribunyaviridae includes eight genera for globally distributed viruses: Orthobunyavirus, Gryffinivirus, Herbevirus, Khurdivirus, Lakivirus, Lambavirus, Pacuvirus, and Shangavirus. Most peribunyaviruses are maintained in vertebrate-arthropod transmission cycles that can include transovarial transmission within arthropods. Others are considered as arthropod-specific viruses that are not capable of infecting vertebrates or have only been associated with vertebrates. Arthropods can be persistently infected. Host and geographic ranges are generally restricted although some peribunyaviruses share common or overlapping ranges. Human infection occurs through blood feeding by a vector arthropod. Infections can result in a diversity of human and veterinary clinical outcomes in a strain-specific manner. Segment reassortment is evident between some peribunyaviruses. Genera are monophyletic based on analysis of the viral L protein and members of a genus share relatively similar genomic organizations and transmission cycles.
Table 1 Peribunyaviridae. Characteristics of members of the family Peribunyaviridae
| Characteristic | Description |
| Example | Bunyamwera virus (BUNV) [S segment: D00353; M segment: M11852; L segment X14383], species Orthobunyavirus bunyamweraense |
| Virion | Enveloped spherical or pleomorphic virions, 80–120 nm in diameter |
| Genome | Three single-stranded, negative sense RNA molecules, S, M, and L, each of about 1 kb, 4 kb, and 6.8 kb, encode the nucleocapsid protein (N; S segment), envelope glycoproteins (Gn and Gc; M segment), and L protein that functions as the RNA-directed RNA polymerase and endonuclease (L; L segment) |
| Replication | Cytoplasmic; primary transcription is primed by “cap snatching” of host cellular RNAs; ribonucleocapsid is involved in primary transcription and unprimed genomic replication |
| Translation | Translation takes place on endoplasmic reticulum bound ribosomes for Gn and Gc and on free ribosomes in the cytoplasm for N and L proteins |
| Host range | Vertebrates and invertebrates (including mammals, birds, mosquitoes, midges, and sandflies) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Polyploviricotina, class Bunyaviricetes, order Elliovirales; 8 genera and 152 species |
Virion
The peribunyavirus particle (Figure 1 Peribunyaviridae) is spherical or pleomorphic, 80–120 nm in diameter (Martin et al., 1985, Marklewitz et al., 2013), and has surface glycoprotein projections of 5–18 nm (Elliott 2014) that are embedded in a lipid bilayer envelope that is about 5 nm thick. The envelope is derived from cellular Golgi membranes. Small, medium, and large viral ribonucleocapsids are 2–2.5 nm in diameter, 200–3,000 nm in length and have helical symmetry.
 |
| Figure 1 Peribunyaviridae. Peribunyavirus particle structure. (Left) Diagrammatic representation of a peribunyavirus virion in cross-section. The surface spikes comprise two glycoproteins termed Gn and Gc. The three helical nucleocapsids are circular and comprise one each of the unique ssRNA segments (L, large; M, medium; S, small) encapsidated by N protein and associated with the L protein. (Middle) Genome organization, transcripts and proteins of Bunyamwera virus. (Right) Negative-stained transmission electron microscopic of California encephalitis virus virions (image CDC/ Dr. Fred Murphy; Dr. Erskine Palmer). |
Physiochemical and physical properties
Information is only available for viruses in the genus Orthobunyavirus. Virions display a density of 1.17–1.19 in sucrose or 1.20 in potassium tartrate; sedimentation coefficient of 350–470S; and a molecular weight of 3–4 × 108 (Bishop et al., 1980).
Proteins
The peribunyavirus virion consists of four structural proteins: two external glycoproteins (Gn and Gc), a nucleocapsid protein (N), and a large protein (L), which possesses RNA-directed RNA polymerase (RdRP) and endonuclease functions (Figure 1 Peribunyaviridae; Table 2 Peribunyaviridae). Non-structural proteins (NSs and NSm) are encoded by some viruses.
Table 2 Peribunyaviridae. Proteins encoded by peribunyaviruses and their approximate masses (kDa)
| Genus | |||||||||
| RNA | Encoded protein | Gryffinivirus | Herbevirus | Khurdivirus | Lakivirus* | Lambavirus | Orthobunyavirus | Pacuvirus | Shangavirus* |
| L segment | L | 260−268 | 282 | 249 | 276 | 252 | 259−263 | 259−261 | 264 |
| M segment | Gn | 31−35 | 35 | 36 | 32 | 55 | 29−41 | 32 | 30 |
| Gc | 61−64 | 56 | 75 | 55 | 67 | 108−120 | 86−88 | 111 | |
| NSm | 11−19 | none | none | none | none | 15−18 | 19 | 27 | |
| S segment | N | 29−30 | 25−27 | 25 | 26 | 26 | 19−26 | 27−28 | 30 |
| NSs | none | none | none | none | none or 10−13 | none | none |
*putative protein masses
Nucleic acid
The peribunyavirus genome includes three, negative-sense RNA segments designated S (small), M (medium), and L (large) (Table 3 Peribunyaviridae). The 5′- and 3′-terminal nucleotide sequences of each segment are highly conserved and complementary among viruses of a given genus. Base-pairing of the terminal nucleotides is predicted to form a stable panhandle structure and non-covalently closed circular RNAs (Raju and Kolakofsky 1989) that form individual ribonucleocapsids when complexed with N and L proteins (Figure 1 Peribunyaviridae). Genomic RNAs are not modified at the 5′- or 3′-ends. Viral mRNAs are truncated relative to the vRNA (Abraham and Pattnaik 1983, Eshita et al., 1985, Marklewitz et al., 2013), are not polyadenylated and possess 5′-methylated caps derived from host cellular mRNAs (Patterson et al., 1984).
Table 3 Peribunyaviridae. RNA segments of typical genus members
| Genus | Virus | RNA segment (bases) | Highly conserved reverse complementary termini | |||
| L | M | S | 3′ | 5′ | ||
| Gryffinivirus | Hedwig virus | 6965 | 4606 | 1079 | ND | ND |
| Herbevirus | Herbert virus | 7428 | 2684 | 1090 | AGTAGTGTGC... | ...GCACACTACT |
| Khurdivirus | Khurdun virus | 6604 | 3161 | 950 | AGTAGTGTACT... | ...AGCACACTACT |
| Lakivirus | Lakamha virus | 7223 | 2623 | 1078 | AGTAGTGTACT... | ...AGCACACTACT |
| Lambavirus | largemouth bass bunyavirus | 6901 | 3570 | 1002 | GTAGTATAC... | ...GCATACTAC |
| Orthobunyavirus | Bunyamwera virus | 6875 | 4458 | 961 | AGTAGTGTACT... | ...AGCACACTACT |
| Pacuvirus | Pacui virus | 6762* | 4386* | 741* | ND | ND |
| Shangavirus | Shuāngào insect virus 1 | 7008* | 4620* | 813* | ND | ND < |
*Open reading frame length; ND: not determined
Carbohydrates
Information is only available for viruses in the genus Orthobunyavirus. Virions may contain asparagine-linked mannose sugars on the Gn and Gc proteins. Carbohydrates make up 7% of orthobunyavirus virion weight (Bishop et al., 1980).
Lipids
Information is only available for viruses in the genus Orthobunyavirus. Virion lipids are derived from the host membranes where viruses mature, and include phospholipids, sterols, fatty acids, and glycolipids. Orthobunyavirus virions contain 20–30% lipids by weight (Bishop et al., 1980).
Genome organization and replication
The general genome organization for viruses of each genus is shown in Figure 2 Peribunyaviridae. The S segment encodes the N and, in the case of some orthobunyaviruses, the NSs proteins in an overlapping reading frame. (Mohamed et al., 2009, Marklewitz et al., 2013, Rodrigues et al., 2014, de Melo et al., 2018). The M segment encodes the Gn and Gc proteins (Fazakerley et al., 1988). Orthobunyaviruses, pacuviruses, and Shuāngào insect virus 1 (the sole member of the genus Shangavirus) also encode an NSm protein between the Gn and Gc proteins ORFs within the polyprotein (Fazakerley et al., 1988, Rodrigues et al., 2014, Li et al., 2015), whereas herbeviruses, khurdiviruses, lambaviruses, and lakiviruses do not encode NSm (Marklewitz et al., 2013, Dimitry Konstantinovich et al., 2015, Kopp et al., 2019, Waltzek et al., 2019). The L segment encodes the L protein (Endres et al., 1989).
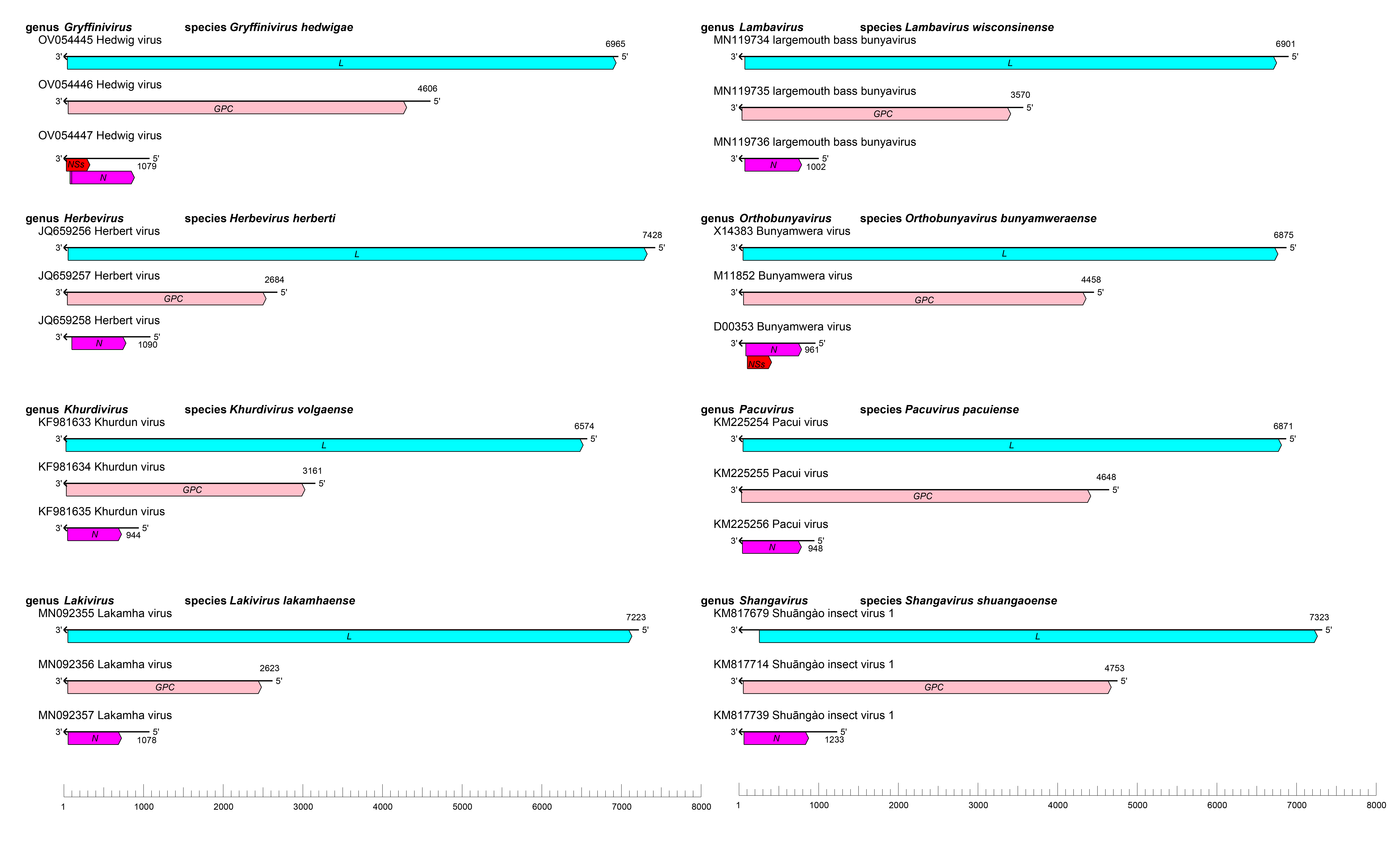 |
| Figure 2 Peribunyaviridae. Generalized coding strategies for the eight genera of peribunyaviruses. vcRNAs are depicted in a 3′→5′ direction and mRNAs are depicted in a 5′→3′ direction. The mRNAs depict ORFs that encode the N, nucleocapsid protein; Gn and Gc, external glycoproteins; L, large protein, as well as the non-structural proteins NSs and NSm. |
Both Gn and Gc proteins are implicated in virion host-cell attachment (Kingsford et al., 1983, Ludwig et al., 1989, Hacker et al., 1995). However, the Gc protein appears to be the primary attachment protein for orthobunyaviruses (Plassmeyer et al., 2005). Following attachment, the virion enters the cell through clathrin-mediated endocytosis and acidification leads to pH-induced insertion of the viral Gc fusion peptide into the endosomal membranes (Jacoby et al., 1993, Plassmeyer et al., 2005, Shi and Elliott 2007). This insertion facilitates the membrane-to-membrane fusion and subsequent release of ribonucleocapsids into the cytoplasm. Only encapsidated RNA segments serve as templates for both primary transcription and genomic replication (Reguera et al., 2013). Complementary 5′- and 3′-termini are promoters for both mRNA and antigenome synthesis (Kohl et al., 2004). The Gn and Gc proteins, and the NSm protein in the case of some viruses, are translated from a single ORF as a polyprotein precursor which is co-translationally cleaved into mature products by a host signal peptidase (Fazakerley and Ross 1989) and the host intramembrane-cleaving protease, signal peptide peptidase (SPP) (Shi et al., 2016). Gn and Gc proteins are modified by N-linked glycans (Madoff and Lenard 1982, Shi et al., 2005) and are targeted to, and retained in the Golgi complex (Lappin et al., 1994) where virions assemble and bud. Virions bud into Golgi cisternae and are transported to the cell surface by the secretory pathway (Salanueva et al., 2003).
Nucleocapsid proteins are abundant in infected cells (Fuller et al., 1983) and encapsidate viral genomic and antigenomic RNA segments, thereby protecting them from degradation. The L protein is responsible for both primary transcription and genomic replication (Elliott and Schmaljohn 2013) and includes a characterized polymerase (RdRP) module shared by all negative-sense RNA viruses within the phylum Negarnavircota (Walter and Barr 2011, Wolf et al., 2018). The L protein N-terminal domain possesses endonuclease activity that cleaves capped oligonucleotides from the 5′-end of host mRNA. This phenomenon is known as “cap snatching”. Subsequently, this host sequence “cap” primes viral mRNA synthesis (Reguera et al., 2010). NSs protein functions have been characterized in orthobunyaviruses as contributing to the shutdown of mammalian, but not mosquito, host cell protein synthesis (Weber et al., 2001, Thomas et al., 2004, Hart et al., 2009). Orthobunyavirus NSs protein has well documented anti-IFN activity in mammalian systems (Weber et al., 2002, Blakqori et al., 2007). As such, NSs is thought to have a role in potentiating the zoonotic capacity of orthobunyaviruses (Hart et al., 2009) by allowing these viruses to overcome the innate immune responses of vertebrate hosts. The Bunyamwera virus NSm protein is a transmembrane protein and targeted to the Golgi complex where budding occurs (Shi et al., 2006), suggesting that NSm may play a role in virion assembly (Fontana et al., 2008). However, NSm may be dispensable for the generation of infectious viral particles (Tilston-Lunel et al., 2015, Kraatz et al., 2018).
Biology
The majority of peribunyaviruses replicate in vertebrates and arthropods and are generally cytopathic in their vertebrate, but not invertebrate, hosts. Others are described as arthropod-specific viruses (Marklewitz et al., 2013) that are not able to replicate in vertebrates. Peribunyaviruses are ecologically diverse and have been found on every continent except Antarctica. A broad diversity of arthropod and vertebrate hosts is evident, as is described on individual genus pages. Peribunyaviruses are associated with a wide range of clinical outcomes (see Genus Orthobunyavirus), ranging from subclinical to fatal. In natural infections of mammals, peribunyaviruses are often targeted to a particular organ or cell type. Genetic reassortment has been demonstrated for many peribunyaviruses both in vitro and in vivo and reassortment has been associated with outbreaks of human disease (see Genus Orthobunyavirus).
Antigenicity
The orthobunyavirus Gn and Gc proteins are responsible for eliciting hemagglutination-inhibiting and neutralizing antibodies (Kingsford et al., 1983). Complement-fixing antibodies are elicited by the nucleocapsid protein.
Derivation of names
Gryffinivirus: named for the fictional character Godric Gryffindor in the Harry Potter novels by J.K. Rowling
Herbevirus: derived from Herbert virus
Lambavirus: derived from largemouth bass bunyavirus
Lakivirus: derived from Lakahma virus
Khurdivirus: derived from Khurdun virus
Orthobunyavirus: derived from Bunyamwera village (Uganda, Africa) where the virus Bunyamwera virus, was first isolated.
Pacuvirus: derived from Pacui virus.
Peribunyaviridae: derived from the Greek περί [perí], meaning around, and the name of the orthobunyavirus, Bunyamwera virus.
Shangavirus: derived from Shuāngào insect virus 1
Genus demarcation criteria
Inclusion in a genus can be defined by:
- Forming a monophyletic clade with other viruses in the genus using the L protein sequence
- Encoding NSs (some orthobunyaviruses) or NSm (orthobunyaviruses, pacuviruses, and shangaviruses)
- N-terminal truncation of the Gc proteins (herbeviruses, lakiviruses)
Relationships within the family
Viruses within the same genus form monophyletic clades based upon phylogenetic reconstruction using the L protein sequence (Figure 3 Peribunyaviridae).
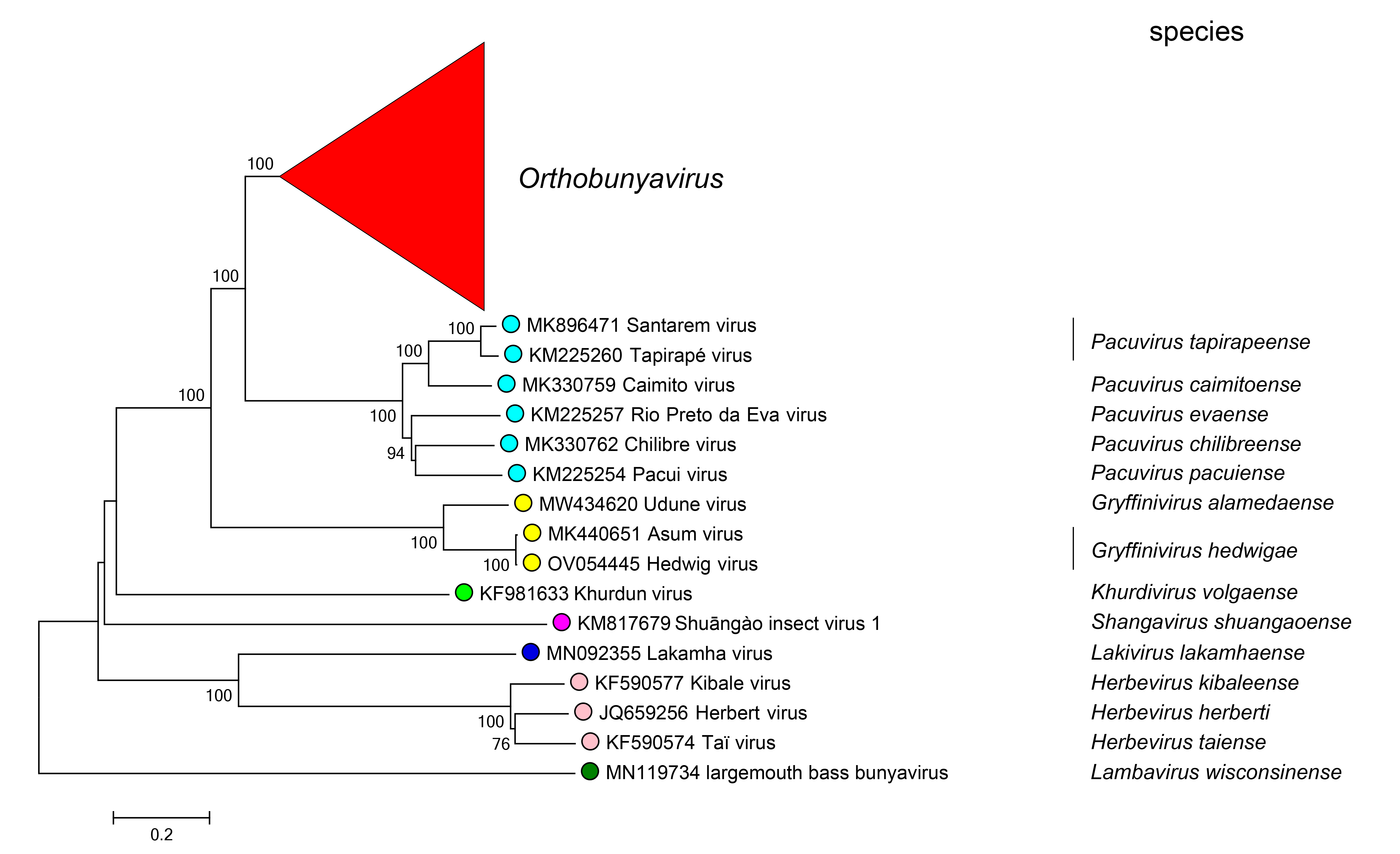 |
| Figure 3 Peribunyaviridae. Phylogenetic analysis of members of the family Peribunyaviridae. The amino acid sequences comprising the L protein of family members were aligned with Clustal W and a neighbor joining tree was produced based on the JTT model using Mega version 7. Numbers at nodes indicate bootstrap support where this was > 70%. Members of the same genus have circles with the same colour: open circles indicate unclassified viruses. A version with the Orthobunyavirus clade expanded is shown on the Orthobunyavirus genus page (Figure 2 Orthobunyavirus). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Viruses of the family Peribunyaviridae are related phylogenetically most closely to those of Fimoviridae and Tospoviridae (Guterres et al., 2017).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Akhtuba virus | L: KF601560; M: KF601561; S: KF601562* | AKTV | |
| Fulton virus | L: MK618652; M: MK618653; S:MK618654* | FULV | (Williams et al., 2019) |
* partial genome
Virus names and virus abbreviations are not official ICTV designations.

