Family: Krittikaviridae
Apoorva Prabhu and Christian Rinke
The citation for this ICTV Report chapter is the summary to be published as
Prabhu, A and Rinke, C. (2026) ICTV Virus Taxonomy Profile: Krittikaviridae 2026, Journal of General Virology, (in press)
Corresponding author: Apoorva Prabhu ([email protected])
Edited by: Mart Krupovic and Evelien Adriaenssens
Posted: February 2026
Summary
The family Krittikaviridae includes dsDNA viruses that are associated with marine archaea in the order Poseidoniales, formerly known as Marine Group II Euryarchaeota (Table 1 Krittikaviridae). They are closely related to other “magroviruses”, a collective term for marine group II viruses. Members of this family have been discovered using metagenomics, and the virions are predicted to contain icosahedral heads and helical tails, these being characteristic of viruses in the class Caudoviricetes. Virus genomes encode genes in both virion morphogenesis (major capsid protein, tail protein) and replication (DNA polymerase B) modules. The family includes the genus Velanvirus and the species Velanvirus brisbanense.
Table 1 Krittikaviridae. Characteristics of members of the family Krittikaviridae
| Characteristic | Description |
| Example | magrovirus_E_01 (PP497039), species Velanvirus brisbanense |
| Virion | Predicted icosahedral capsid and helical tail |
| Genome | Linear dsDNA of 80 kbp |
| Replication | Virus-encoded family B DNA polymerase |
| Translation | Unknown |
| Host range | Archaea (predicted) |
| Taxonomy | Realm Duplodnaviria, kingdom Heunggongvirae, phylum Uroviricota, class Caudoviricetes, order Magrovirales: genus Velanvirus, species Velanvirus brisbanense |
Virion
Morphology
Members of this family, like many other archaeal viruses, have been recovered using metagenomics (Nishimura et al., 2017, Philosof et al., 2017, Xu et al., 2023, Zhou et al., 2023, Prabhu et al., 2025). The complete genome of magrovirus_E_01 (species Velanvirus brisbanense) encodes a virion morphogenetic module, including genes for head and tail proteins, such as the HK97-fold major capsid protein, that are typical of viruses in the class Caudoviricetes (Figure 1 Krittikaviridae). Other morphogenesis proteins include prohead protease, portal protein, tail tape measure protein, and the large subunit of the terminase. In addition to these proteins, krittikavirids share relatively close sequence and gene content similarity with tailed haloviruses and are therefore predicted to have icosahedral heads and helical tails (Baquero et al., 2020, Liu et al., 2021).
Nucleic acid
The complete genome of magrovirus_E_01 (species Velanvirus brisbanense) has been assembled as a linear contig of 79,954 bp and is predicted to encode 88 proteins. The genes comprising the morphogenetic module and DNA replication and repair modules are predicted to be transcribed in the same direction (Figure 1 Krittikaviridae).
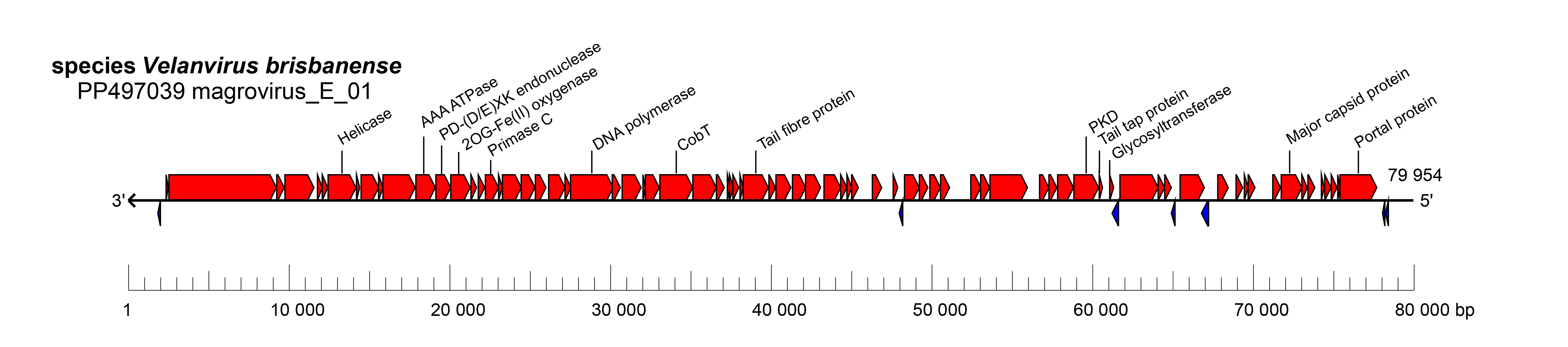 |
| Figure 1 Krittikaviridae. Genome organisation of magrovirus_E_01, a member of the family Krittikaviridae. Boxes indicate open reading frames as annotated on GenBank accession PP497039. Note that not all annotations are shown in the figure. |
Genome organization and replication
Similar to other magroviruses, krittikavirid genomes encode genes for DNA replication and repair, such as DNA polymerase and primase.
Biology
Krittikavirids have been discovered using metagenomics and have not yet been cultured. The lack of integrase and transposase proteins suggests a lytic lifestyle (Figure 1 Krittikaviridae). Other than the morphogenetic and replication modules, krittikavirid genomes encode various auxiliary proteins, such as chaperonin GroEL for proper protein folding, CobT, wWA domain protein potentially involved in cellular adhesion, a PKD-domain containing protein, a protein with a helix-turn-helix motif, and a GT4 glycosyltransferase (Figure 1 Krittikaviridae).
Derivation of names
Krittikaviridae: from Krittika, a legendary princess bathing in an Indian river (Ganga) according to ancient Indian mythology.
Velanvirus: from Velan, the child of Krittika, who was born and flowed in the waters, according to ancient Indian mythology
Demarcation criteria for taxa within the family
There is currently only one monotypic genus in the family.
Relationships within the family
The family includes the single species Velanvirus brisbanense (Figure 2 Krittikaviridae). Comparative genomics with unclassified viruses in IMG/VR and from Brisbane River Estuary dataset (PRJNA1024631) reveals a cluster of viruses sharing 20–40% of their genes, and these may represent additional genera.
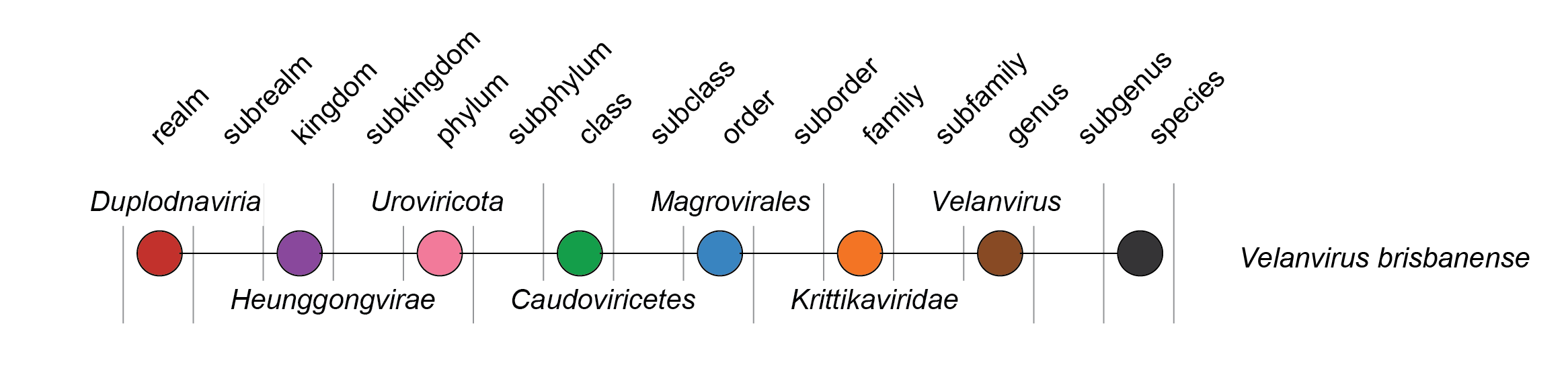 |
| Figure 2 Krittikaviridae. Taxonomy of the family Krittikaviridae. |
Relationships with other taxa
The family shares < 60% of proteins with its nearest relative, according to the demarcation criterion set forward for archaeal viruses (Liu et al., 2021).
Members of the order Magrovirales show close similarities with viruses in the family Druskaviridae (class Caudoviricetes), which infect hyperhalophilic archaea (Zhou et al., 2023). Magroviruses can be divided into multiple clades: clade B comprises viruses in the family Aoguangviridae, clade A comprises viruses in the family Apasviridae, and clade E comprises viruses in the family Krittikaviridae. It is possible that viruses in clades C, D and X could be members of additional families but complete circular genomes are not yet available.

