Family: Turriviridae
Sydnie K. Chase and Jamie C. Snyder
The citation for this ICTV Report chapter is the summary published as Chase & Snyder (2024):
ICTV Virus Taxonomy Profile: Turriviridae 2024, Journal of General Virology (2024) 105: 002000
Corresponding author: Jamie C. Snyder (jcsnyder@cpp.edu)
Edited by: Mart Krupovic and Evelien Adriaenssens
Posted: May 2024
Summary
The family Turriviridae includes viruses with a dsDNA genome of 16–17 kbp. Virions are spherical with a diameter of about 75 nm, and comprise a host-derived internal lipid membrane surrounded by a proteinaceous icosahedral capsid shell (Table 1 Turriviridae). Members of the family Turriviridae infect archaea of the genera Sulfolobus and Saccharolobus, which thrive in extreme environments. In the case of Sulfolobus turreted icosahedral virus 1, viral infection results in cell lysis and the virus genome does not integrate into that of the host.
Table 1 Turriviridae. Characteristics of members of the family Turriviridae
| Characteristic | Description |
| Example | Sulfolobus turreted icosahedral virus 1 (AY569307), species Alphaturrivirus yellowstonense, genus Alphaturrivirus |
| Virion | Turreted icosahedral particle (about 75 nm in diameter) with an internal membrane layer |
| Genome | Circular, dsDNA genome of 16–17 kbp |
| Replication | Lytic (Sulfolobus turreted icosahedral virus 1) or temperate |
| Translation | Not described |
| Host range | Hyperthermophilc and acidophilic archaea of the genera Saccharolobus and Sulfolobus |
| Taxonomy | Realm Varidnaviria, kingdom Abadenavirae, phylum Produgelaviricota, class Belvinaviricetes, order Belfryvirales: one genus and two species |
Virion
Morphology
Virions of viruses in the family Turriviridae are morphologically similar to sphaerolipoviruses, the main difference being the structure of the major capsid protein (MCP) which contains a double jelly roll fold (Krupovic et al., 2018). Virions are spherical (75 nm in diameter) with turret-like protrusions extending about 12 nm above the capsid shell. The virions consist of an external proteinaceous capsid shell based on a pseudo T=31d icosahedral symmetry and an internal host-derived lipid membrane (Maaty et al., 2006, Happonen et al., 2010, Veesler et al., 2013) (Figure 1 Turriviridae). Encased in this spherical virion is a circular dsDNA genome. Two members of the family, Sulfolobus turreted icosahedral virus 1 (STIV1) and Sulfolobus turreted icosahedral virus 2 (STIV2) are nearly identical in morphology yet behave slightly differently (Overton et al., 2023). Virions of STIV1 have been shown in bind to the pili of the host which are likely to serve as the primary receptor (Munson-McGee et al., 2018, Hartman et al., 2019).
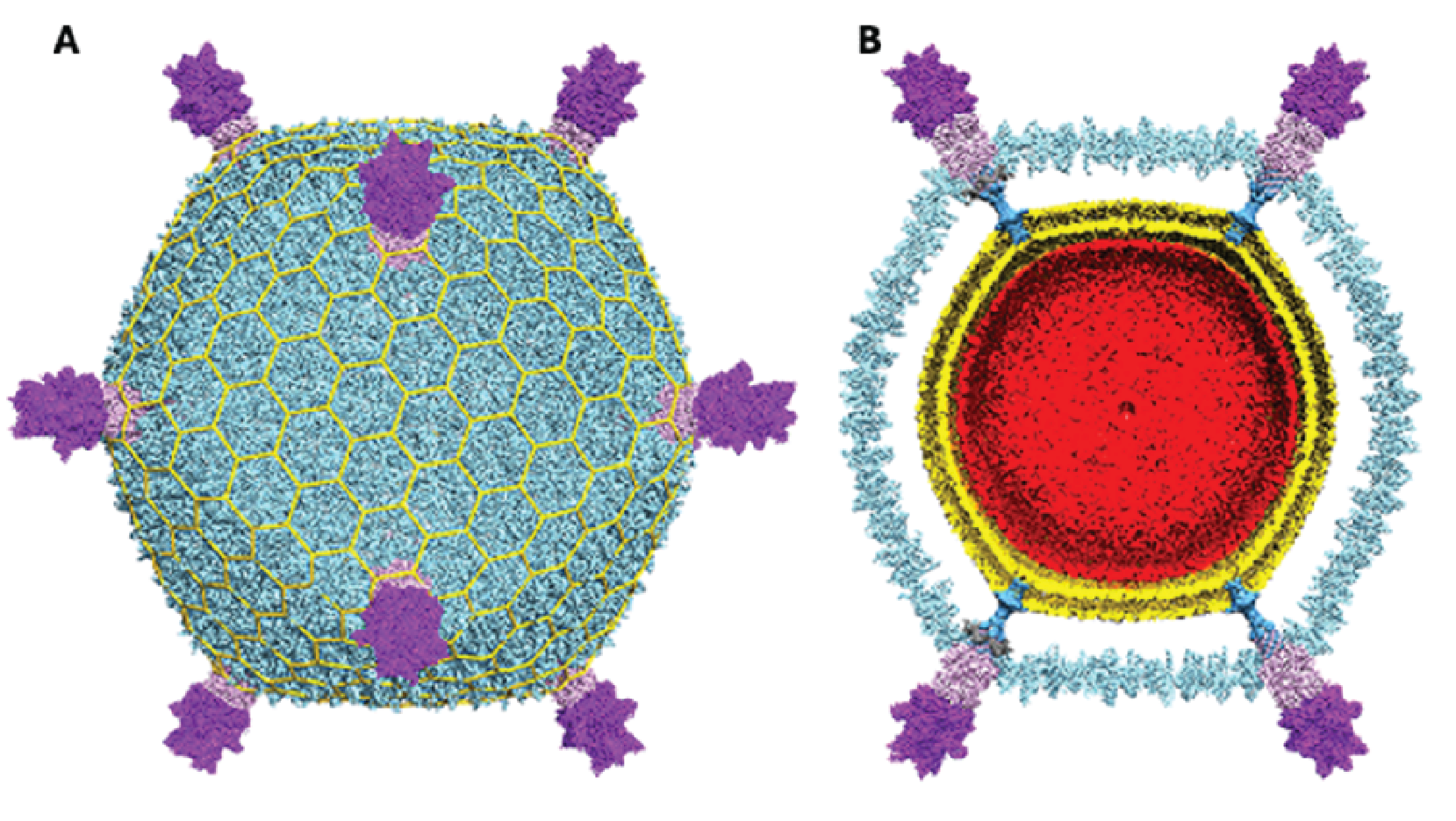 |
| Figure 1 Turriviridae. Cryo-EM reconstruction of Sulfolobus turreted icosahedral virus 1. (A) Virus reconstruction with turrets at five-fold points of symmetry. (B) Cross-section revealing the internal lipid layer (gold) and the dsDNA genome (red). Adapted from (Overton et al., 2023). |
Physicochemical and physical properties
Virion buoyant density in cesium sulfate is about 1.236 g/cm3 (Rice et al., 2004). Virions are highly heat stable.
Nucleic acid
Virions contain a circular dsDNA genome comprising of 16–17 kbp for STIV1 and STIV2.
Proteins
The major capsid protein (MCP, B345) forms the outer proteinaceous capsid shell of turriviruses and contains the double jelly roll fold (Krupovic et al., 2018). This double jelly roll fold is seen in viruses infecting the eukaryotic and bacterial domains and is evidence for a common viral ancestry (Overton et al., 2023). The turret structures that extend from each of the 12 fivefold vertices comprise a C381 pentamer stacked on top of the A223 pentamer (Figure 2 Turriviridae). The A223 pentamer is bound to A55, which is anchored in the internal lipid envelope. A223 and C381 are also composed of jelly roll domains, like the MCP (B345). The N-terminal jelly roll of the A223 pentamer interfaces with the capsid shell, where the N-terminal ![]() -strands of the first jelly roll of A223 interdigitate with
-strands of the first jelly roll of A223 interdigitate with ![]() -strands of A55, forming a 10-stranded antiparallel, hemolysin-like
-strands of A55, forming a 10-stranded antiparallel, hemolysin-like ![]() -barrel. This structure forms the penton base. The second jelly roll of A223 interfaces with the N-terminus of C381 and together these proteins form a triple jelly roll. The A223 and A55 interactions anchor the capsid shell to the internal lipid membrane (Overton et al., 2023).
-barrel. This structure forms the penton base. The second jelly roll of A223 interfaces with the N-terminus of C381 and together these proteins form a triple jelly roll. The A223 and A55 interactions anchor the capsid shell to the internal lipid membrane (Overton et al., 2023).
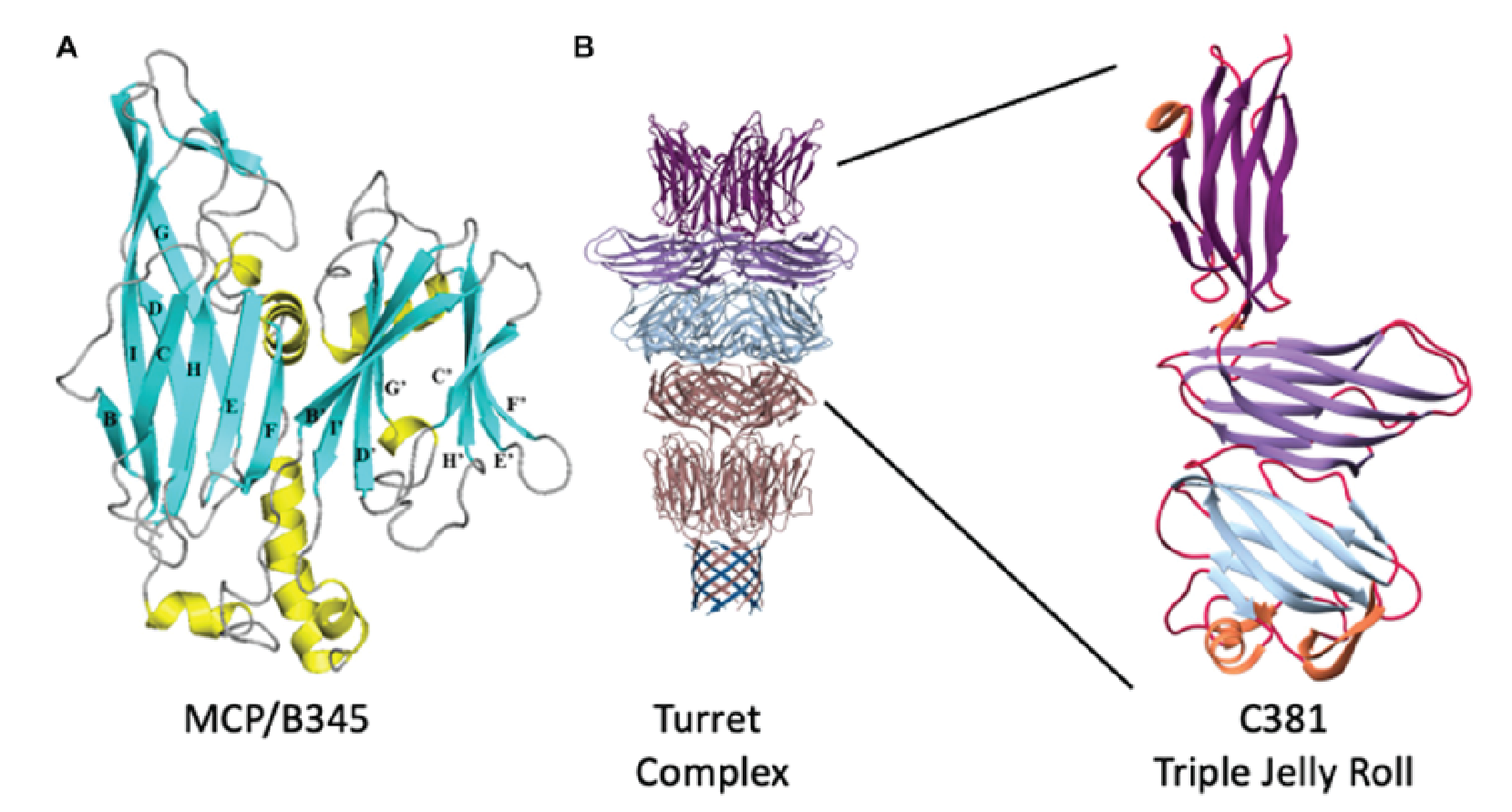 |
| Figure 2 Turriviridae. Structural proteins of Sulfolobus turreted icosahedral virus 1. (A) Ribbon representation of B345 (MCP). Blue ribbons are |
Lipids
Virions contains a subset of host-derived lipids for inner membrane formation.
Genome organization and replication
The genome of STIV1 has 38 confirmed CDSs while that of STIV2 has 34 predicted CDSs. In STIV1, all but three confirmed CDSs are on one strand (Figure 3 Turriviridae), and in the case of STIV2, all but six are on one strand. Viral replication is lytic in the case of STIV1, whereas the life cycle of STIV2 remains poorly understood and the virus can no longer be propagated in culture (Overton et al., 2023). STIV1 uses an unusual mechanism of cell lysis, which relies on the formation of pyramidal structures on the cell surface through which the virions are eventually released. In particular, STIV1 protein C92 produces 7-sided pyramid structures in the host lipid membrane that open during cell lysis like the petals of a flower to allow mature virions to escape (Snyder et al., 2011, Snyder et al., 2013) (Figure 4 Turriviridae). The pyramid-dependent virion release has been also described for archaeal viruses from three other unrelated families (Bize et al., 2009, Wang et al., 2018, Baquero et al., 2021). Notably, however, STIV2 does not encode a homolog of the STIV1 C92 and the mechanism of its release remains unclear (Happonen et al., 2010). Although STIV1 and STIV2 do not encode integrases, some unclassified members of the family Turriviridae have been shown be integrated in the genomes of S. acidocaldarius species (Anderson et al., 2017, Mao and Grogan 2017), indicating that turriviruses can be either lytic or temperate.
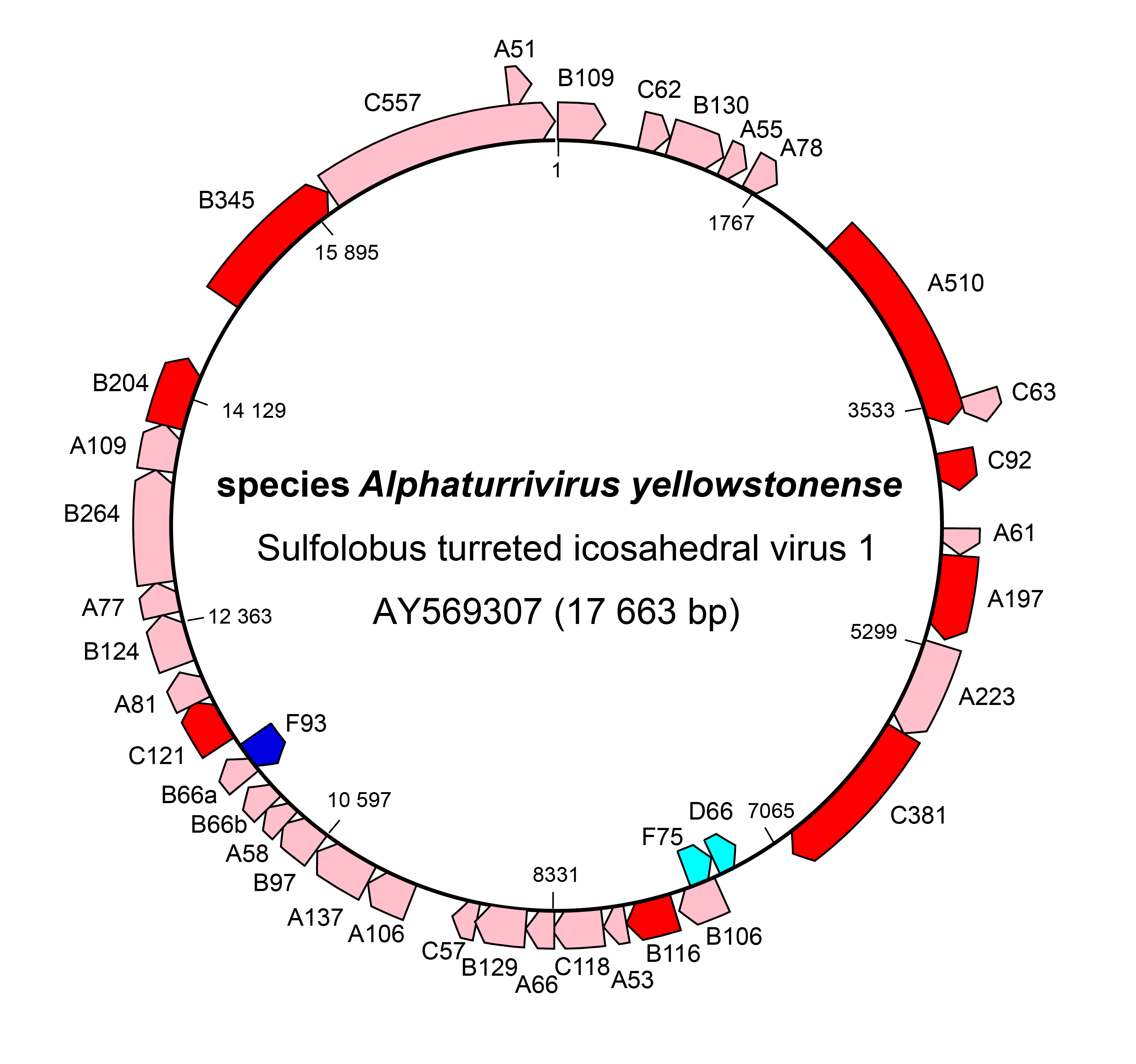 |
| Figure 3 Turriviridae. Genome organization of Sulfolobus turreted icosahedral virus 1. Dark red and dark blue: CDSs for which genetic or structural information is known. |
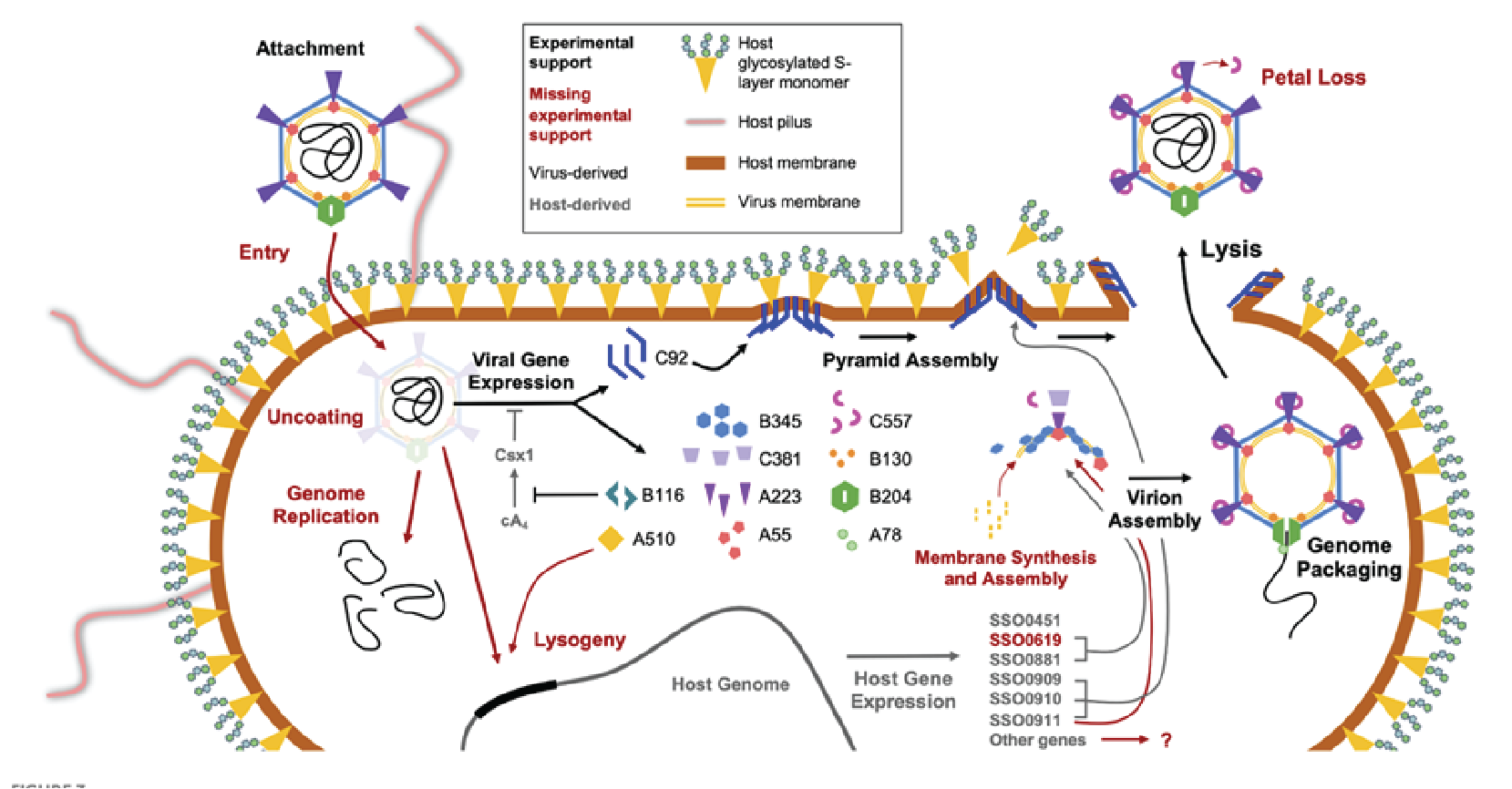 |
| Figure 4 Turriviridae. Replication cycle of Sulfolobus turreted icosahedral virus 1. The replication cycle starts with viral attachment (top left) and ends with viral lysis (top right). Figure reproduced from (Overton et al., 2023). |
Biology
Viruses in the family Turriviridae infect archaea in the genera Sulfolobus and Saccharolobus. No known archaeal viruses or their hosts are known to infect eukaryotic or bacterial domains (Wirth and Young 2020). Like many archaeal viruses, turriviruses can be collected from most extreme natural environments, such as acidic hot springs and areas with volcanic activity, which are almost completely populated by archaeal viruses and their hosts (Munson-McGee et al., 2018).
Derivation of names
Turriviridae: from the Latin turris, for turret
hveragerdiense: from Hverakjalki, the valley in Iceland where STIV2 was isolated
yellowstonense: from Yellowstone Park where STIV1 was first isolated.
Relationships within the family
Members of the family share overall virion organization, sequence similarity and gene synteny. Genome comparisons between STIV1 and STIV2 exhibit high homology across most of the genome (25/34 CDSs). Nucleotide identities exceed 70% in some CDSs (Overton et al., 2023).
Relationships with other taxa
Structural studies of turrivirus MCPs reveal a double jelly roll fold (Maaty et al., 2006). The same fold is also found in a wide range of eukaryotic viruses (e.g., members of the family Adenoviridae and the phylum Nucleocytoviricota), bacteriophages (e.g., members of the families Tectiviridae and Corticoviridae) as well as archaeal viruses of the families Chaacviridae (Laso-Pérez et al., 2023) and Skuldviridae (Medvedeva et al., 2022), which infect ANME-1 and Asgard archaea, respectively. Genetically, there are only four CDSs in STIV1 that have recognizable homologs in other viruses, these being members of the families Rudiviridae and Lipothrixviridae.

