Family: Thaspiviridae
Jong-Geol Kim, Khaled S. Gazi, Mart Krupovic and Sung-Keun Rhee
The citation for this ICTV Report chapter is the summary published as Kim et al., (2021):
ICTV Virus Taxonomy Profile: Thaspiviridae 2021 J Gen Virol. 2021 102(7):001631.
Corresponding author: Sung-Keun Rhee ([email protected])
Edited by: Mart Krupovic and Stuart G. Siddell
Posted: June 2021
Summary
Members of the family Thaspiviridae have linear dsDNA genomes of 27 to 29 kbp (Table 1. Thaspiviridae). The family includes one genus, Nitmarvirus, with one species, Nitmarvirus NSV1. Thaspiviruses infect members of the genus Nitrosopumilus (phylum Thaumarchaeota), a group of ammonia-oxidizing archaea widespread in moderate environments, where they play important roles in nitrification and carbon cycling. Accordingly, viruses of the family Thaspiviridae are mesophilic and neutrophilic. Although viral replication leads to inhibition of host growth, accompanied by severe reduction in the ammonia oxidation rate, virus propagation does not lead to degradation of the host chromosome or decrease in host cell counts. Members of the family Thaspiviridae display spindle-shaped morphology with short fibers at one pole, thereby resembling archaeal viruses of the families Fuselloviridae and Halspiviridae.
Table 1. Thaspiviridae. Characteristics of members of the family Thaspiviridae
| Characteristic | Description |
| Example | Nitrosopumilus spindle-shaped virus 1 (MK570053), species Nitmarvirus maris |
| Virion | Spindle-shaped virion, measuring 64 ± 3 nm in diameter and 112 ± 6 nm in length, with short fibers at one pole |
| Genome | Linear dsDNA (27−29 kbp) with 176 bp terminal inverted repeats |
| Replication | Non-lytic, chronic infection. Protein-primed family B DNA polymerase is involved in viral DNA replication |
| Translation | Not characterized |
| Host range | Ammonia-oxidizing archaea of the genus Nitrosopumilus |
| Taxonomy | Single genus with one species |
Virion
Morphology
Virions of Nitrosopumilus spindle-shaped virus 1 (NSV1) are 64±3 nm in diameter and 112±6 nm in length, with a short tail at one pole (Figure 1. Thaspiviridae, left). This morphology is similar to that of members of the families Fuselloviridae (Krupovic et al., 2014) and Halspiviridae (Bath et al., 2006). A large fraction of virions produced by infected cells remain attached to the cell surface (Figure 1. Thaspiviridae, right). A fraction of cell-associated virions display elongated morphology (Kim et al., 2019). Similar plasticity of spindle-shaped virion morphology is observed for other spindle-shaped viruses of the family Bicaudaviridae; namely, Acidianus two-tailed virus (ATV) (Häring et al., 2005) and Sulfolobus monocaudavirus 1 (SMV1) (Uldahl et al., 2016). Changes in virion morphology can occur outside of the host cells.
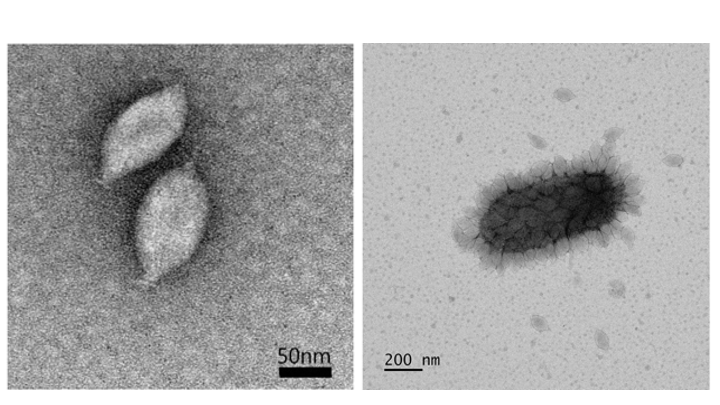 |
| Figure 1. Thaspiviridae. (left) Transmission electron micrograph of negatively-stained virions of Nitrosopumilus spindle-shaped virus 1. Scale bar: 50 nm. (right) Virus particles attached to the surface of a host cell. Scale bar, 200 nm |
Physicochemical and physical properties
Virions of NSV1 are stable at ambient seawater conditions and are tolerant to environmental variations, including a wide range of pH (pH 3−9), salinities (0.1%−20%), and temperatures (up to 55°C).
Nucleic acid
Three isolates of Nitrosopumilus spindle-shaped virus 1 (NSV1-1, NSV1-2 and NSV1-3) have genomes of linear dsDNA of approximately 27 kbp, 29 kbp and 27 kbp, respectively. They all have inverted terminal repeats of 176 bp. The GC content of these genomes is 29.8% (Kim et al., 2019). The virus genomic DNA might be covalently bound to a terminal protein required for the postulated protein-primed genome replication.
Proteins
Nano LC-MS/MS analysis reveals the presence of 10 virus-encoded proteins in purified NSV1 virions, including the 81 aa putative major capsid protein which has two highly hydrophobic α-helical regions, a feature typical of the major capsid proteins of fuselloviruses and halspiviruses (Krupovic et al., 2014).
Lipids
None reported.
Carbohydrates
None reported. The virus encodes two glycosyltransferases which could be involved in glycosylation of virion proteins, as is the case for many other archaeal viruses.
Genome organization and replication
Members of the family Thaspiviridae have linear dsDNA genomes, similar to viruses of the family Halspiviridae (Bath and Dyall-Smith 1998). The three NSV1 isolates NSV1-1, NSV1-2, and NSV1-3) are >95% identical in nucleotide sequence, and have 48, 51, 48 open reading frames, respectively, and are considered as isolates of the same species, Nitmarvirus NSV1. No appreciable sequence similarity is detected between the proteins encoded in the genome of NSV1 and those of other known archaeal or bacterial viruses, except for the protein-primed family B DNA polymerase, which is one of nine proteins that display similarity to proteins encoded by various marine Thaumarchaeota and bacteria. The proteins showing similarity to thaumarchaeotal proteins include the protein-primed family B DNA polymerase (ORF3), proliferating cell nuclear antigen (PCNA; ORF44), two glycosyltransferases (ORF21 and ORF40) and a DNA-binding protein (ORF45) (Table 2.Thaspiviridae).
Table 2. Thaspiviridae. Core genes of NSV1 of the family Thaspiviridae.
| ORFa | Annotation |
| 3 | Protein primed family B DNA polymerase |
| 4 | Possible terminal protein |
| 13 | Putative major capsid protein |
| 16 | AAA+ ATPase, Cdc6-like |
| 19 | DNA binding protein with winged helix-turn-helix domain |
| 21 | Glycosyltransferase |
| 34 | DNA N-6-adenine methyltransferase |
| 40 | Glycosyltransferase |
| 44 | Proliferating cell nuclear antigen (PCNA) |
| 45 | DNA binding protein with ribbon-helix-helix domain |
aORF designations are based on their positions within the genome of NSV1 (MK570053).
Viral DNA replication occurs concurrently with host cell growth and enables continuous virion production and release through a non-lytic mechanism (Kim et al., 2019). Replication of the NSV1 genome likely occurs by a mechanism similar to that used by eukaryotic adenoviruses (Miralles et al., 1989), whereby a virus-encoded protein-primed family B DNA polymerase uses a terminal protein to prime the DNA synthesis at the terminal inverted repeats. In addition, the virus encodes a proliferating cell nuclear antigen (PCNA) which is also likely to be involved in virus genome replication.
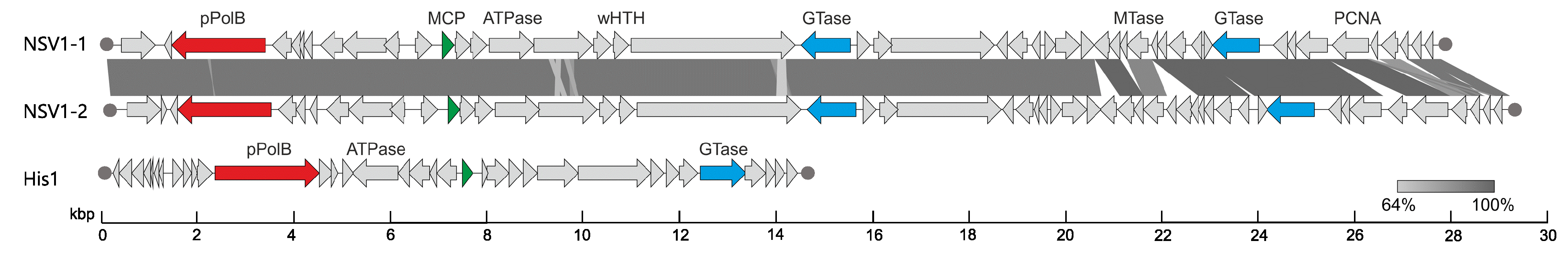 |
| Figure 2. Thaspiviridae. Genome maps of Nitrosopumilus spindle-shaped virus 1 isolates NSV1-1 and NSV1-2, and of the haloarchaeal halspivirus His1. Shared ORFs are connected by shaded areas based on sequence identity. Functionally equivalent (but not necessarily homologous) genes are indicated with matching colors. Filled circles indicate terminal inverted repeats. Abbreviations: pPolB, protein-primed family B DNA polymerase; MCP, major capsid protein (putative); wHTH, winged helix-turn-helix; GTase, glycosyltransferase; MTase, DNA methyltransferase; PCNA, proliferating cell nuclear antigen. |
Biology
Host cells of the genus Nitrosopumilus in the phylum Thaumarchaeota are widespread and abundant in marine ecosystems (Karner et al., 2001). They play key roles in nitrification by mediating ammonia oxidation. The host range of NSV1 is very narrow possibly reflecting the lack of cultured representatives of Nitrosopumilus. NSV1-like particles are frequently observed in marine environments, especially in particle-rich seawaters and marine sediments, indicating a cosmopolitan distribution in marine ecosystems (Kim et al., 2019). NSV1 displays rapid adsorption kinetics, with > 60% of virus progeny being bound to host cells (Kim et al., 2019).
Previously characterized archaeal viruses infect extremophilic hosts and often possess unique virion morphologies not observed among bacterial or eukaryotic viruses (Prangishvili et al., 2017), suggesting that these archaea-specific morphotypes have evolved to adapt to extreme environments. However, that fact that NSV1 infects mesophilic marine Thaumarchaeota means that these unique archaeal virus morphotypes are also associated with mesophilic archaea. The broad distribution of spindle-shaped viruses suggests a deep evolutionary history of this virus group in the archaeal domain, probably dating back to the last archaeal common ancestor (Krupovic et al., 2020).
The non-lytic infection strategy seen with members of the family Thaspiviridae, suggests that the virus is well adapted to efficiently infect its chemolithoautotrophic ammonia-oxidizing archaeal hosts in the resource-poor ocean environment. It is clear that NSV1 severely affects the metabolism of infected thaumarchaeal host cells (Kim et al., 2019), with a potential impact on global carbon and nitrogen cycling.
Antigenicity
Not known
Derivation of names
Nitmarvirus: from Nitrosopumilus-infecting marine virus
Thaspiviridae: from thaumarchaeal and spindle-shaped virus
Relationships within the family
Three NSV1 viruses have been isolated. Their genome sequences are >95% identical to each other and they are considered to all be members of the species Nitmarvirus NSV1.
Relationships with other taxa
Virion organization, genome architecture and the postulated genome replication mechanism of thaspiviruses are most similar to those of members of the family Halspiviridae (Bath and Dyall-Smith 1998) (Figure 2. Thaspiviridae). Nevertheless, viruses from the two families lack detectable sequence similarity and they infect hosts belonging to different phyla of archaea (Thaumarchaeota and Euryarchaeota, respectively). The genomes of members of the family Thaspiviridae do not display appreciable sequence similarity to other known viruses.

