Subfamily: Comovirinae
Genus: Comovirus
Distinguishing features
Comoviruses have bipartite genomes encapsidated by two capsid proteins (CP); these viruses are transmitted by beetles.
Virion
The comovirus capsid is made of two types of polypeptides (large CP: 40–45 kDa and small CP: 21–27 kDa). The small CP suppresses RNA silencing and surface-exposed amino acids are required for this function.
Genome organisation and replication
The 5ʹ- and 3ʹ-UTRs of RNA1 and RNA2 are similar in sequence but not identical. RNA2 is translated into two largely overlapping polyproteins that are processed into three domains. Production of the smaller polyprotein results from internal initiation at a downstream AUG, which is placed in a more favourable context than the upstream AUG (Figure 1 Comovirus). The 58K protein released from the N-terminus of the larger polyprotein (P2) is necessary for replication of RNA2. The 48K protein released from the N-terminus of the smaller polyprotein (P2ʹ) is the movement protein (MP), with a typical LxxPxL motif. The CP domains are encoded at the C-terminus of both polyproteins. The MP and the CPs are required for cell-to-cell movement of the virus (Wellink and Van Kammen 1989). The MP is a structural component of tubular structures containing virus-like particles that traverse the cell wall. The C-terminal region of the MP also interacts with the large CP (Carvalho et al., 2003). RNA1 is translated into a single polyprotein that is processed into five domains, through alternative processing pathways (Figure 1 Comovirus). The N-terminal 32K protein limits the processing of the RNA1-encoded polyprotein in cis and assists the processing of the RNA2-encoded polyprotein (Peters et al., 1992). This protein is often referred to as the protease co-factor or Co-Pro. The replication block on the RNA1-encoded polyprotein includes the 58K protein with sequence motifs characteristic of an NTP-binding helicase, the VPg, the Pro and the Pol. The 32K Co-Pro and 58K NTB proteins are involved in inducing the cytopathic structure through proliferation of ER-derived membranes (Carette et al., 2002).
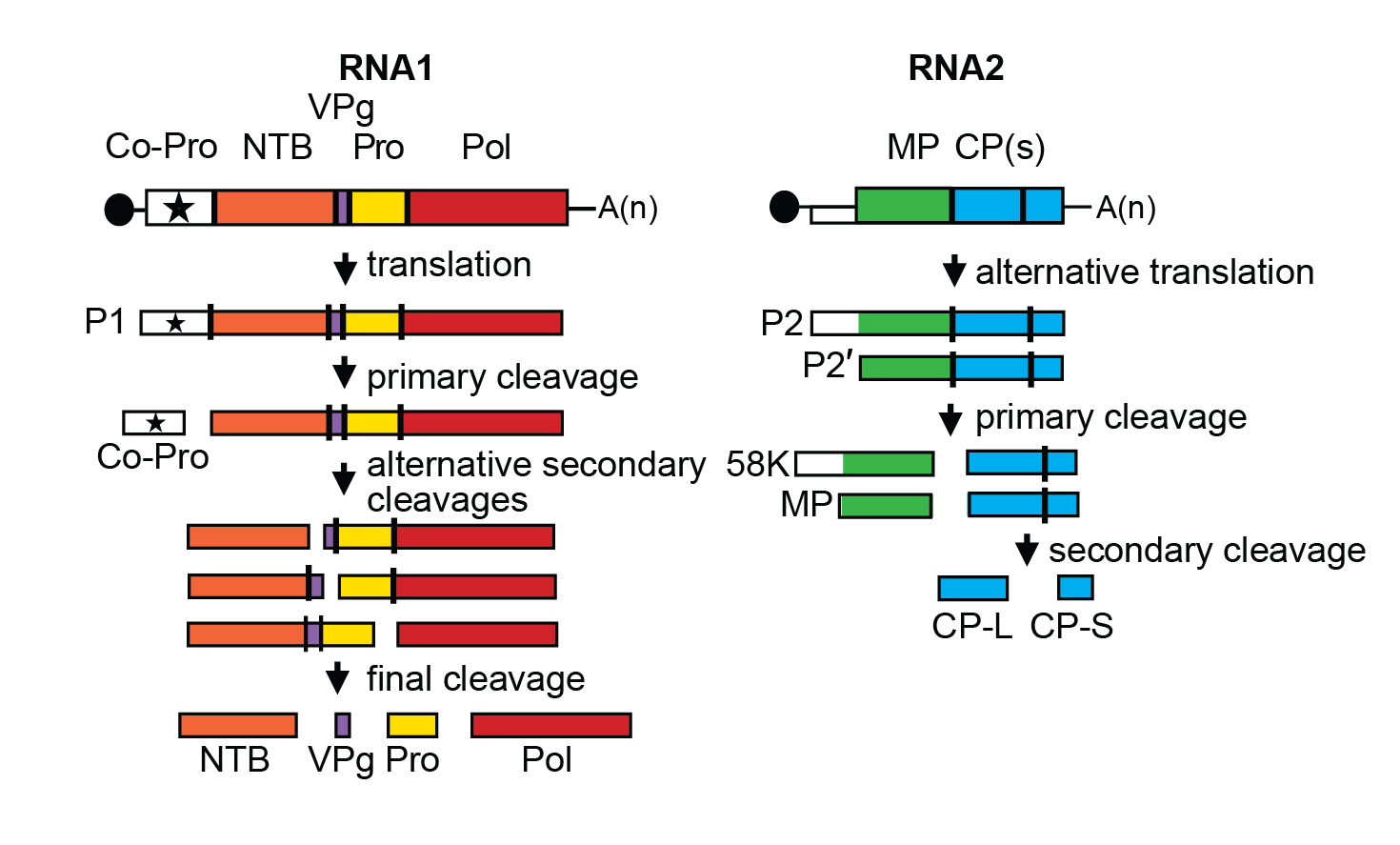 |
| Figure 1 Comovirus. Genome organization and polyprotein processing of cowpea mosaic virus. The ORFs are boxed and the function of the proteins is indicated. MP: movement protein; CPL and CPS: large and small capsid proteins; Co-Pro: proteinase co-factor; NTB: NTP-binding proteins; Pro: proteinase; Pol: RNA-dependent RNA polymerase. Proteolytic cleavage sites are indicated on the polyproteins with the vertical lines. All intermediate and final cleavage products have been detected in infected cells. The black circles at the 5′-end of the RNA represents the VPg, and A(n) at the 3′-end the poly-A tail. |
Biology
Comoviruses have narrow host ranges, 11 of the 15 species being restricted to a few species of the family Leguminosae. Mosaic and mottle symptoms are characteristic, but usually not ringspots. Transmission in nature is exclusively by beetles, especially members of the family Chrysomelidae. Beetles retain their ability to transmit virus for days or weeks.
Species demarcation criteria
See discussion under family description.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| turnip ringspot virus | RNA-1: FJ516745; RNA-2: FJ516746 | TuRSV |
Virus names and virus abbreviations are not official ICTV designations.
Turnip ringspot virus (TuRSV) is related to radish mosaic virus (RaMV). They infect similar hosts. The degree of aa sequence identity between the two viruses is close to the proposed species demarcation criteria (73% aa sequence identity in the combined CP region and 80% aa sequence identity in the Pro-Pol region) (Petrzik and Koloniuk 2010). It is not known whether re-assortment between the RNAs of TuRSV and RaMV is possible. Therefore, the taxonomic position of TuRSV as a distinct species in the genus Comovirus or as a distant strain of the species Radish mosaic virus remains unclear.

