Family: Nairoviridae
Genus: Orthonairovirus
Distinguishing features
There are 51 distinct viruses currently assigned to the genus Orthonairovirus. These viruses typically infect specific argasid or ixodid ticks, which transmit them to small vertebrates (birds and mammals) that in turn then server as sources for tick infection. Some orthonairoviruses are transmitted transovarially in ticks.
Virion
Morphology
Orthonairovirions are spherical in shape, 80–120 nm in diameter, and surrounded by a membrane envelope that is decorated with glycoprotein (GP) spikes composed of GN and GC subunits (Figure 1 Orthonairovirus). Isolated ribonucleoprotein (RNP) complexes are composed of individual segment genomic RNA encapsidated by nucleoprotein (N). Hazara virus (HAZV), kupe virus (KUPEV), Erve virus (ERVEV), and Crimean-Congo hemorrhagic fever virus (CCHFV) Ns are structurally conserved in the head domain (Korolev et al., 1976, Tantawi et al., 1980, Munz et al., 1981, Booth et al., 1991, Rwambo et al., 1996, Surtees et al., 2015, Wang et al., 2015).
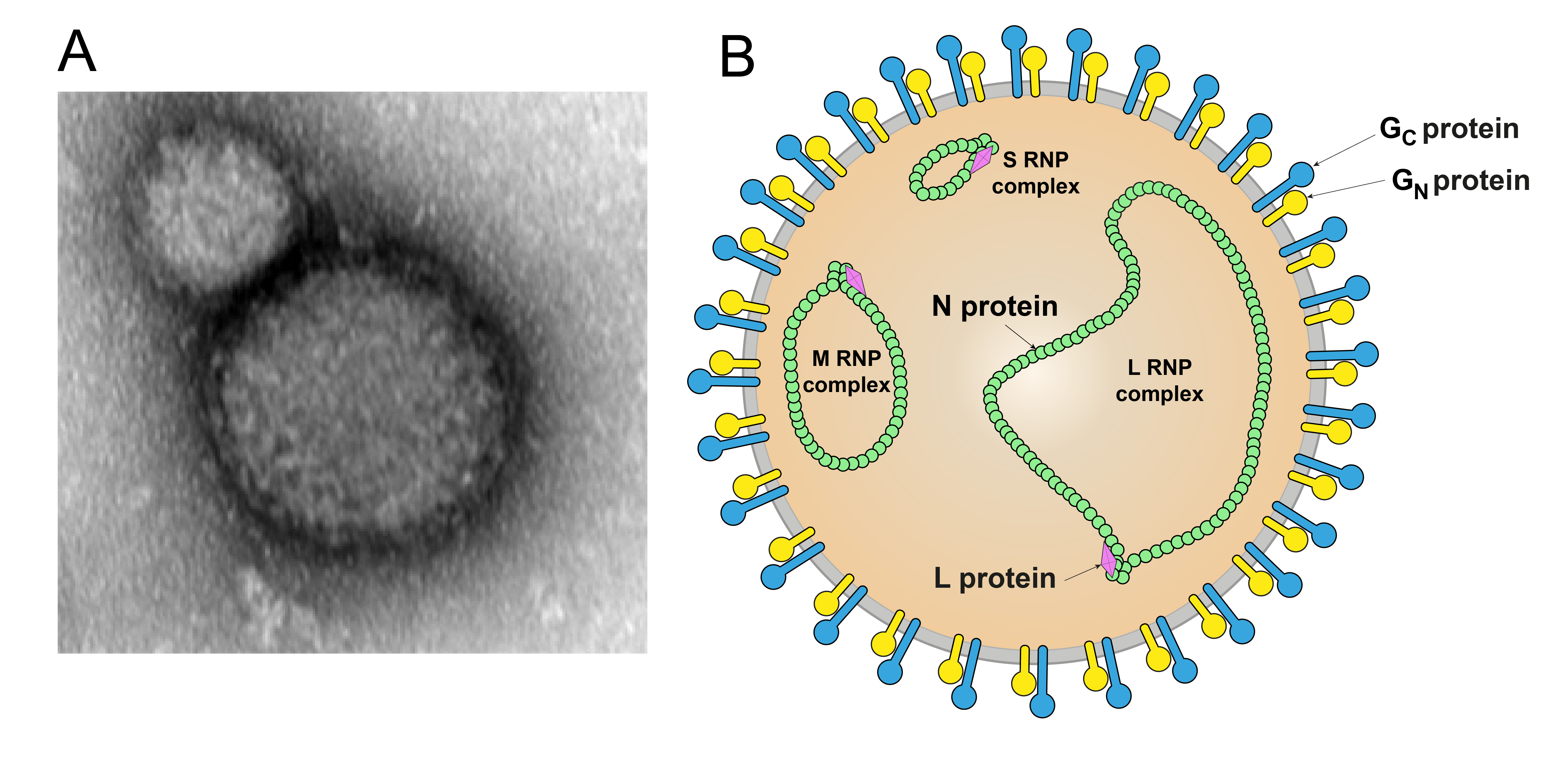 |
| Figure 1 Orthonairovirus. A) Electron micrograph of a Crimean-Congo hemorrhagic fever virus particle. Magnification 50 000× (Courtesy of Mateja Poljšak-Prijatelj and Marko Kolenc, Institute of Microbiology and Immunology, Ljubljana, Slovenia). B) Schematic illustration of an orthonairovirus particle. Shown is the spherical and enveloped (grey) particle with glycoprotein spikes (GN yellow, GC blue) inserted in a bilaminar lipid envelope. The S (small), M (medium), and L (large) RNP (ribonucleoprotein) complexes inside the particle consist of N (nucleoprotein, green) and L (large protein, pink). |
Physicochemical and physical properties
The virion Mr is 300×106 to 400×106 and has an S20,W of 350–500. Virion buoyant densities in sucrose and CsCl are 1.16–2.0 and 1.20–1.21 g cm−3, respectively. Virions are sensitive to heat, lipid solvents, detergents, and formaldehyde (Donets et al., 1977, Terpstra 1983).
Nucleic acid
Orthonairovirus genomes are linear trisegmented RNA molecules of negative polarity. The genomes are about 17.2–20.1 kb (small [S] segment: 1.4–3.8 kb; medium [M] segment: 3.9–5.9 kb; large [L] segment: 11.7–12.6 kb) (Obijeski and Murphy 1977, Clerx et al., 1981, Kuhn et al., 2016, Alkhovsky et al., 2017, Kapuscinski et al., 2021). The Mr of the genome ranges from 4.8×106–8×106 and accounts for 1–2% of the weight of the virion (Bishop et al., 1980, Clerx et al., 1981).
Proteins
Orthonairoviruses typically express three structural proteins (Table 1 Orthonairovirus). The most abundant structural protein in an orthonairovirion is N, which encapsidates the orthonairoviral genomic segments. The least abundant protein is L, which mediates viral genome replication and transcription. Two glycoprotein subunits, GN and GC, mediate virion entry. GN and GC and non-structural proteins (encoded by some, but not all orthonairoviruses (Freitas et al., 2020)) are encoded as a precursor polyprotein.
Table 1 Orthonairovirus. Location and function of orthonairovirus structural proteins.
| Protein | Location, mass, and function | Reference |
| Nucleoprotein (N) | Structural virion protein (60–68 kD). Oligomerizes and encapsidates orthonairoviral genomic segments and hence component of RNPs. Functions as an endonuclease. Facilitates cell-to-cell spread by modulating tight junction claudin 1. Antagonizes type I interferon production by RIG1 ubiqutinylation inhibition | (Carter et al., 2012, Guo et al., 2012, Wang et al., 2012, Devignot et al., 2015, Surtees et al., 2015, Wang et al., 2015, Hawman and Feldmann 2023, Ohta et al., 2023) |
| Glycoprotein (GP) | Structural virion protein consisting of two subunits (GN: 30–45 kD, GC: 72–84 kD). Produced via proteolytic cleavage from the genome-encoded precursor GPC (which may also result in production of nonstructural proteins, such as GP38, GP85, GP160, and NSm in the case of CCHFV). Inserts into virion membranes as trimeric GP spikes composed of GN and GC. As a class II fusion machine, GP mediates virion cell-surface and internal receptor binding, virion-cell membrane fusion and, thereby cell entry. | (Sanchez et al., 2006, Altamura et al., 2007, Bergeron et al., 2007, Bergeron et al., 2015, Mishra et al., 2020, Hulswit et al., 2021, Li et al., 2022, Mishra et al., 2022, Bost et al., 2023) |
| Large protein (L) | Structural virion protein (250–450 kD) with RdRP, helicase, and endoribonuclease domains. Component of the RNP inside virions. Oligomerizes and mediates transcription and replication of viral RNA segments. Contains an ovarian tumor family-like domain (vOTU) that is conserved among all orthonairoviruses and suppresses type I interferon responses and deISGylase activity by broadly removing ISG15-conjugated proteins. Mediates cap-snatching for viral mRNA capping. | (Scholte et al., 2017, Dzimianski et al., 2019, Mirza et al., 2019, Devignot et al., 2020, Tchesnokov et al., 2020, Pyle et al., 2021) |
Genome organization and replication
The orthonairovirus genome consists of three negative-sense RNA molecules, termed S (small), M (medium), and L (large) (Figure 2 Orthonairovirus).
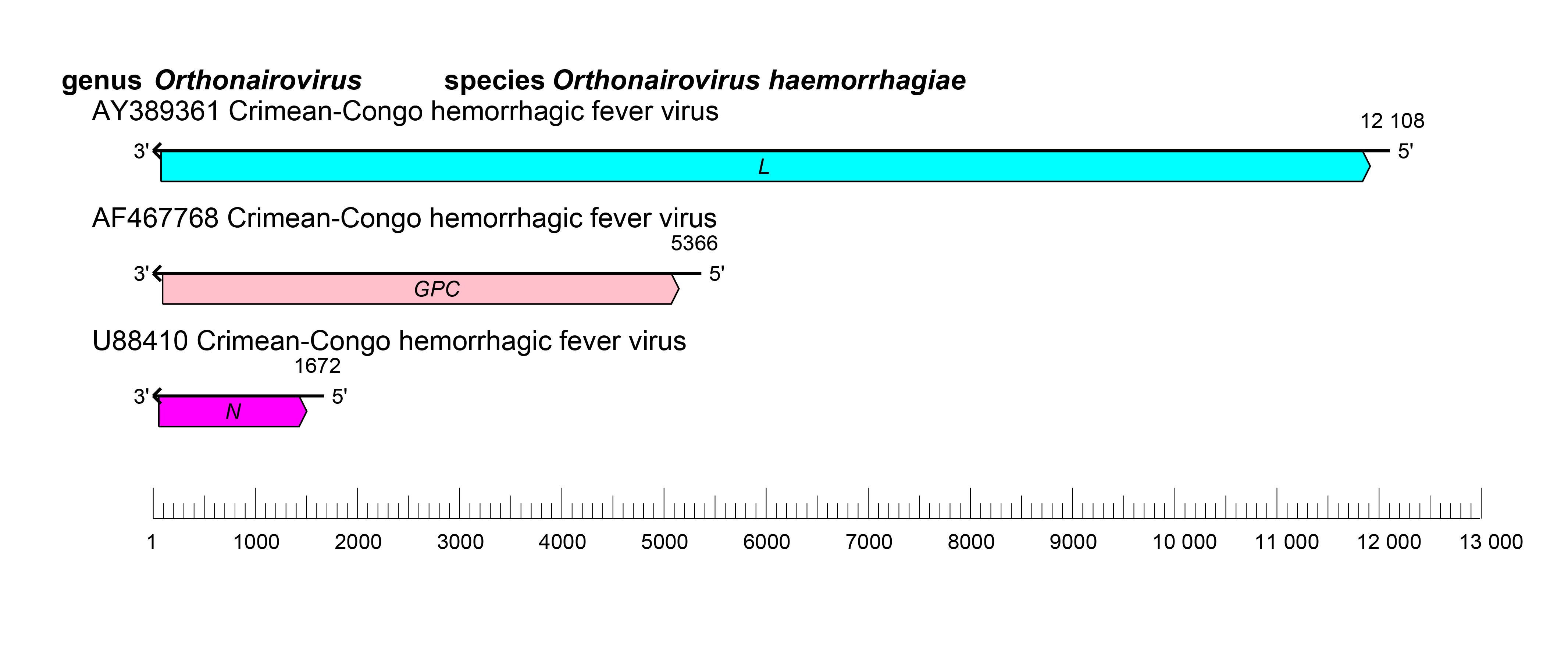 |
| Figure 2 Orthonairovirus. Schematic representation of orthonairovirus genome organization. |
Orthonairovirus genomic segments assume circular forms via non-covalent binding of complementary and highly conserved 3′-and 5′-terminal sequences (11 nt at the extreme ends). The consensus terminal nucleotide sequences of the S, M, and L segments are typically characterized by a genus-specific 5′-genomic segment terminus 5′-UCUCAAAGA-3′ and 3′-genomic segment terminus of 5′-UCUUUGAGA-3′. The nairovirid, and hence orthonairovirus, promoters are unique among bunyavirals. They are bipartite and are located within the complementary terminal segment regions that include these 11 nts (promoter element 1 [PE1]) and adjacent segment-specific nucleotide stretched (PE2) (Matsumoto et al., 2019, Mega et al., 2020, Neriya et al., 2022). Genomic segment reassortment occurs among viruses belonging to the same species but not among viruses of different species.
Orthonairovirus infection starts with virion attachment, mediated by GN and GC, to unknown cell-surface receptors and entry via the endosomal route (Simon et al., 2009, Garrison et al., 2013, Shtanko et al., 2014) (Figure 3 Orthonairovirus).
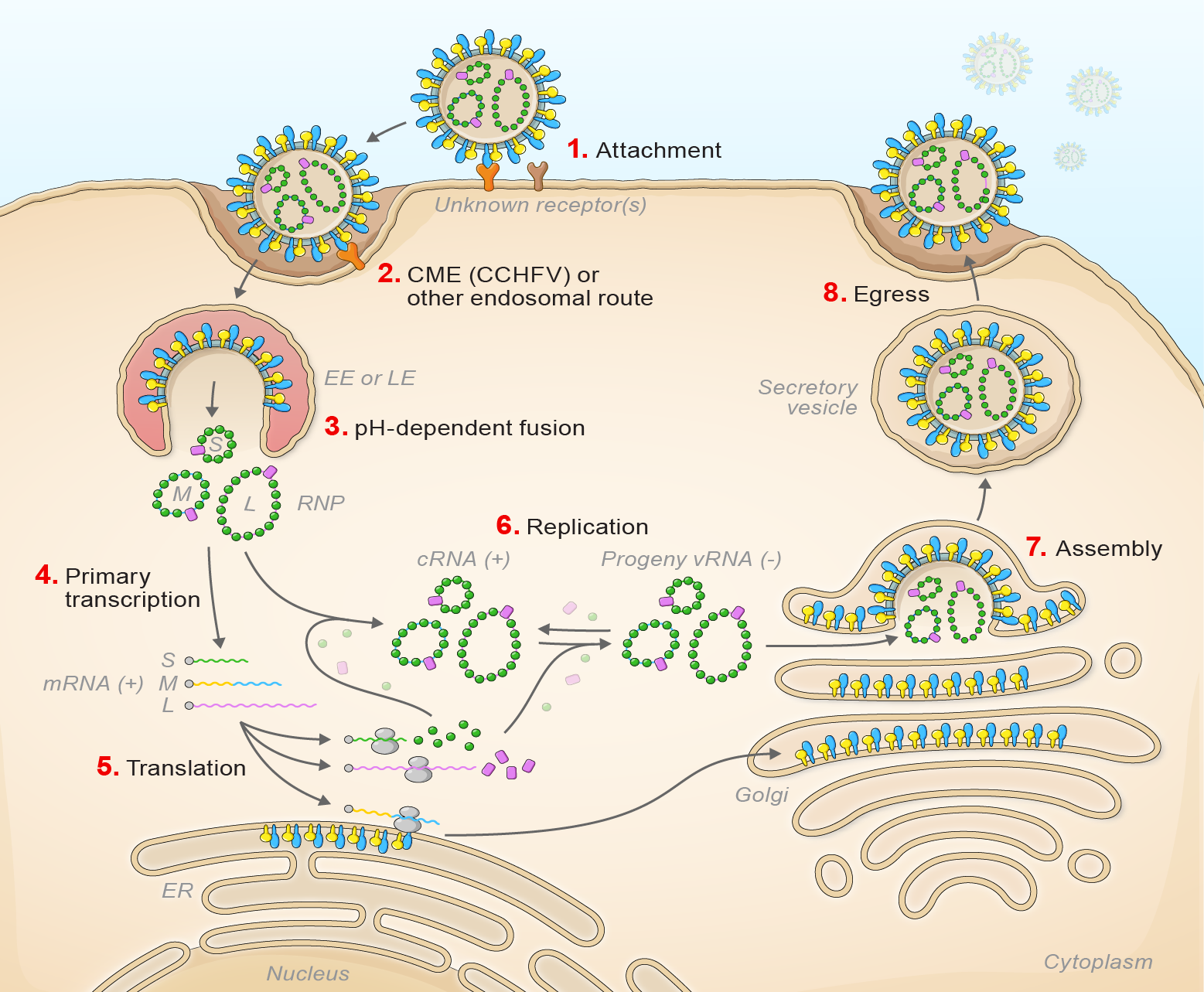 |
| Figure 3 Orthonairovirus. Lifecycle of orthonairoviruses. (1) Virion attachment; (2) virion uptake; (3) virion-cell membrane fusion; (4) transcription; (5) translation; (6) replication; (7) virion assembly; and (8) virion egress. |
Viral fusion with the host cell results in early or late endosomal release, depending on the virus, of the virion RNP complex into the cytoplasm. This pH-dependent fusion event likely requires the previous participation of an intracellular receptor (Garrison et al., 2013, Shtanko et al., 2014). During primary transcription L generates uncapped antigenomic RNA that are then capped using host-cell derived capped primers (cap snatching) (Holm et al., 2018). L and S segment-transcribed mRNAs are translated by free ribosomes. M segment-transcribed mRNA is translated by membrane-bound ribosomes, with the expressed GPC co-translationally and post-translationally cleaved by cellular proteases and glycosylated by cellular glycosidases to yield GN and GC and sometimes non-structural glycoproteins. The synthesis of the antigenomic by L protein serves as a template for genomic RNA replication. Secondary transcription amplifies the synthesis of mRNAs and genome replication. During morphogenesis, GN and GC accumulate in the Golgi apparatus, modified host membranes are acquired, and virions bud into the Golgi cisternae (Booth et al., 1991, Rwambo et al., 1996, Vincent et al., 2003, Sanchez et al., 2006, Bergeron et al., 2015).
Biology
Most orthonairoviruses are capable of alternately replicating in arthropods (argasid or ixodid ticks) and vertebrates (alcid, larid, or struthionid birds; hipposiderid, molossid, pteropodid, and verspertillionid bats; soricid eulipotyphla; lagomorphs; murid rodents, pangolins (Pholidota); or ungulates), and cause little or no cytopathogenicity in their hosts (Spengler et al., 2016). Transovarial, transstadial, and venereal transmission have been demonstrated for CCHFV. In general, distinct orthonairoviruses have a strictly limited range of arthropod vectors and occupy a specific ecological niche. The range of vertebrate hosts is mainly determined by the ecology of their vectors. The most important human orthonairovirus with public-health impact is CCHFV, which is tick-borne and endemic in much of Asia, Africa, and Southern and Eastern Europe, where it causes often severe and frequently fatal viral hemorrhagic fever (Pigott et al., 2017, Blair et al., 2019, Belobo et al., 2021, Temur et al., 2021, Fereidouni et al., 2023, Hawman and Feldmann 2023). Other viruses causing occasional disease in humans are Aigai virus (AIGV; febrile disease, viral hemorrhagic fever) (Midilli et al., 2009, Salehi-Vaziri et al., 2016, Papa et al., 2022), Dugbe virus (DUGV; febrile disease) (Georges et al., 1980, Burt et al., 1996), ERVEV (thunderclap headache) (Treib et al., 1998), Issyk-kul virus (ISKV; febrile disease) (L'Vov D et al., 1980, L'Vov D et al., 1984a), Kasokero virus (KASV; febrile disease) (Kalunda et al., 1986), Nairobi sheep disease virus (NSDV [discouraged synonym: Ganjam virus]; febrile illness) (Dandawate et al., 1969, Rao et al., 1981, Baron and Holzer 2015), Soldado virus (SOLV; febrile disease) (Chastel et al., author's transl, Chastel et al., 1981b), Sōnglǐng virus (SGLV; febrile disease) (Ma et al., 2021), Tǎchéng tick virus 1 (febrile disease) (Liu et al., 2020, Zhang et al., 2021), Tamdy virus (L'Vov D et al., 1984b, Lvov 1994, Moming et al., 2021), and Yezo virus (YEZV; febrile disease) (Rao et al., 1981, Kodama et al., 2021). The most significant orthonairovirus with veterinary importance is NSDV, which is also tick-borne and causes lethal hemorrhagic gastroenteritis in small ruminants in Africa and India (Baron and Holzer 2015, Krasteva et al., 2020). Another orthonairovirus of veterinary importance is Soldado virus (SOLV), which causes lethal infections in juvenile marine birds (Converse et al., 1975b, Chastel et al., author's transl, Chastel et al., 1981b, Chastel et al., 1990).
Antigenicity
One or both of the envelope glycoprotein subunits have hemagglutinating and neutralizing antigenic determinants. Complement-fixing antigenic determinants are principally associated with the nucleoprotein (Shope 1985, Elliott 1990, Bertolotti-Ciarlet et al., 2005).
Species demarcation criteria
Members of different species in the genus are <93% identity in L amino-acid sequence.
Phylogenetic relationships
Phylogenetic relationships across the genus have been estimated using maximum likelihood trees generated from complete N, GPC, and L amino-acid sequences (Figure 2 Nairoviridae).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference | |
| Ahun virus | (Dacheux et al., 2014) | |||
| Āntú virus | S: OQ207701; M: OQ207702; L: OQ207703 | ANTV | (Li et al., 2023) | |
| Bakel virus | BAKV | |||
| cencurut virus | S: OM645182; M: OM645181; L: OM645180 | CENV | (Low et al., 2023) | |
| Elliðaey virus | (Moss et al., 1986) | |||
| Finch Creek virus | S: OK493379; M: OK493378; L: OK493377 | FCV, FINCV | (Major et al., 2009, O'Brien et al., 2022) | |
| Foula virus | (Nuttall et al., 1986) | |||
| Fraser Point virus | FPV | |||
| Garm virus | (Lvov et al., 2015) | |||
| Grímsey virus | (Moss et al., 1986) | |||
| Gubbo nairovirus | S: OP804628; M: OP804625; L: OP514653 | GUBV | (Ortiz-Baez et al., 2023) | |
| Inner Farne Island virus | (Nuttall et al., 1986) | |||
| Island of May virus | (Nuttall et al., 1986) | |||
| Kachemak Bay virus | KBV | (Ritter and Feltz 1974) | ||
| Kao Shuan virus | KSV | (Doherty et al., 1976) | ||
| lamgora virus | S: LC671782; M: LC671781; L: LC671780* | LMGV | (Ozeki et al., 2022) | |
| lamusara virus | S: LC671714; M: LC671713; L: LC671712 | LMSV | (Ozeki et al., 2022) | |
| Méihuā Mountain virus | S: ON184086; M: ON184085; L: ON184084 | MHMV | (Zhang et al., 2022) | |
| Mykines virus | (Nuttall et al., 1986) | |||
| Nàyǔn tick nairovirus | S: KP141755* | NTNV | ||
| Omo virus | OMOV | (Rodhain et al., 1985) | ||
| orthonairovirus sp. isolate YS | S: MW391929; M: MW391928; L: MW391927 | |||
| pangolin orthonairovirus | S: ON024089; M: ON024088; L: ON024087 | PONV | ||
| Paramushir virus | S: MH124636; M: MH124635; L: MH124634 | PARV | (Lvov et al., 1976) | |
| Pathum Thani virus | PTHV | (Bishop et al., 1980) | ||
| Pretoria virus | PREV | (Converse et al., 1975a) | ||
| Puffin Island virus | PIV | (Gould et al., 1983) | ||
| Rondônia virus | S: MN560621; M: MN560623; L: MN560626* | (Blomström et al., 2019) | ||
| Wǔfēng Crocidura attenuata orthonairovirus 1 | S: OM030323; M: OM030322; L: OM030321 | WfCAV1 | - |
Virus names and virus abbreviations are not official ICTV designations.
*Coding region sequence incomplete.

