Family: Nairoviridae
Jens H. Kuhn, Sergey V. Alkhovsky [Альховский Сергей Владимирович], Tatjana Avšič-Županc, Éric Bergeron, Felicity Burt, Koray Ergünay, Aura R. Garrison, Marco Marklewitz, Ali Mirazimi, Anna Papa [Άννα Παπά], Janusz T. Pawęska, Jessica R. Spengler and Gustavo Palacios
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Gustavo Palacios (gustavo.palacios@mssm.edu)
Edited by: Holly Hughes, Jens H. Kuhn, Stuart G. Siddell and Peter J. Walker
Posted: May 2019, updated September 2020, March 2024
Summary
Nairoviridae is a family for negative-sense RNA viruses with genomes of about 17.2–21.1 kb (Table 1 Nairoviridae). The family includes seven genera (Norwavirus, Ocetevirus, Orthonairovirus, Sabavirus, Shaspivirus, Striwavirus, and Xinspivirus). These viruses are maintained in and/or transmitted by arthropods among birds and mammals. Norwaviruses and orthonairoviruses can infect humans, causing mild, severe, and sometimes-fatal diseases. Nairovirids produce enveloped virions containing two or three single-stranded RNA segments with open reading frames (ORFs) that encode a nucleoprotein (N), sometimes a glycoprotein precursor (GPC), and large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain.
Table 1 Nairoviridae. Characteristics of members of the family Nairoviridae.
| Characteristic | Description* |
| Example | Crimean-Congo hemorrhagic fever virus (S segment: U88410; M segment: AF467768; L segment: AY389361), species Orthonairovirus haemorrhagiae |
| Virion | Enveloped, spherical virions 80–120 nm in diameter with heterodimeric surface spikes |
| Genome | Two or three single-stranded RNA molecules (segments): small (S; 1.4–5.5 kb), medium (M; 3.3–5.9 kb), and large (L; 9.1–14.9 kb) |
| Replication | Ribonucleoprotein (RNP) complexes containing anti-genomic RNA serve as coding templates for synthesis of genomic RNA |
| Translation | Proteins are produced from capped and non-polyadenylated mRNAs. The 5′-cap structure is obtained via cap-snatching from cellular mRNAs |
| Host range | Arthropods (norwaviruses, oceteviruses, orthonairoviruses, sabaviruses, shaspiviruses, striwaviruses, xinspiviruses), birds (orthonairoviruses), and mammals (norwaviruses, orthonairoviruses) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Polyploviricotina, class Bunyaviricetes, order Hareavirales; the family includes eight genera and 60 species |
* mostly based on experiments with mammalian orthonairovirids
Viruses assigned to each of the seven genera form a monophyletic clade based on phylogenetic analysis of L protein/RdRP and NP sequences. Viruses from all seven genera share one or more of the following characteristics: (i) enveloped spherical virions; (ii) bisegmented or trisegmented negative-sense RNA genome without polyadenylated tracts at the 3′-end; (iii) genomic sequence complementarity at the 5′- and 3′-ends; and (iv) capped but not polyadenylated virus mRNAs.
Avian Hosts
Genus Orthonairovirus. Some orthonairoviruses infect alcid or larid seabirds, or ostriches (Struthionidae). Orthonairovirus genomes are trisegmented.
Arthropod Hosts
Genus Norwavirus. Běijí nairovirus (BJNV) and Grotenhout virus (GRHV)infect ixodid ticks. BJNV also may infect humans. Norwavirus genomes are bisegmented.
Genus Ocetevirus. Red goblin roach virus 1 (RGRV1) infects ectobiid cockroaches. Ocetevirus genomes are trisegmented.
Genus Orthonairovirus. Almost all members of the genus have been found argasid and/or ixodid ticks.
Genus Sabavirus. South Bay virus (SBV) infects ixodid ticks. Sabavirus genomes are bisegmented.
Genus Shaspivirus. Shāyáng spider virus 1 (SySV1) infects araneid spiders. Shaspivirus genomes are trisegmented.
Genus Striwavirus.Sānxiá water strider virus 1 (SxWSV1) infects water striders. Striwavirus genomes are trisegmented.
Genus Xinspivirus.Xīnzhōu spider virus (XSV) infects araneid spiders. Xinspivirus genomes are trisegmented.
Mammalian Hosts
Genus Orthonairovirus. Many viruses assigned to this genus infect mammals through transmission by argasid or ixodid ticks. Mammalian hosts include hipposiderid, molossid, pteropodid, and verspertillionid bats, soricid eulipotyphla, lagomorphs, murid rodents, pangolins (Pholidota), and ungulates.
Virion
Morphology
Only known for members of genera Norwavirus and Orthonairovirus (see Norwavirus and Orthonairovirus genus pages).
Physicochemical and physical properties
Only known for members of the genus Orthonairovirus (see Orthonairovirus genus page).
Nucleic acid
Nairovirids have bisegmented (S and L segments) or trisegmented (S, M, and L segments) negative-sense RNA genomes (Clerx et al., 1981). Viral mRNAs are not polyadenylated and contain a 5′ methylated cap and 10–18 non-templated nucleotides at the 5′-end that are derived from host cell mRNAs via cap snatching. Further information is only known for members of the genus Orthonairovirus (see Orthonairovirus genus page).
Proteins
Nairovirids typically express three structural proteins. The most abundant structural protein in a nairovirion is N, which encapsidates the nairoviral genomic segments. The least abundant protein is L, which mediates viral genome replication and transcription. Two glycoprotein subunits, GN and GC, are produced from GPC and mediate virion entry of trisegmented nairovirids. Specifics are only known for members of the genus Orthonairovirus (see Orthonairovirus genus page).
Genome organization and replication
S RNA encodes N, M RNA encodes GPC, and the L RNA encodes L with its RdRP, helicase, and endonuclease domains (Figure 1 Nairoviridae). Specifics are only known for members of the genus Orthonairovirus (see Orthonairovirus genus page).
 |
| Figure 1 Nairoviridae. Genome organization of representative nairovirids. ORFs are colored according to the predicted protein function (L, L protein gene; GPC, glycoprotein precursor gene; N, nucleoprotein gene). |
Biology
Most nairovirids have been found in arthropods (predominantly in argasid and ixodid ticks, but also in ectobiid cockroaches, staphylinid beetles, araneid and thomisid spiders, and water striders). Only norwaviruses and orthonairoviruses are known to cause diseases (see Norwavirus and Orthonairovirus genus pages).
Antigenicity
Systematic antigenicity studies have only been reported for orthonairoviruses (see Orthonairovirus genus page).
Derivation of names
abuhammadense: after أبو حماد (Abū Ḥammād), Egypt
abuminaense: after أبو مينا (Abū Mīnā), Egypt
amblyommae: after the host genus name Amblyomma
americanum: after the Latin americanus, meaning of America
aranei: after the Latin araneus, meaning spider
artashatense: after Արտաշատ (Artashat), Armenia
australiaense: after Australia
avalonense: after Avalon Peninsula, Canada.
bandiaense: after Bandia Forest, Senegal
beijisense: after Běijí (北极镇), Chin
buranaense: after Бурана (Burana), Kyrgyzstan
bushkeyense: after Bush Key, USA
chimense: after Chimgon, Uzbekista
clomorense: after Clo Mor, Scotland, UK
dermacentoris: after the host genus name Dermacentor
dugbeense: after Dugbe, Nigeria
erveense: after the Erve river, France
esteroense: after Estero Real, Cuba
gossasense: after Gossas, Senegal
grotenhoutense: after Grotenhout, Belgium
haemorrhagiae: after the Latin haemorrhagia, meaning “hemorrhage”
hazaraense: after ہزارہ (Hazara) District, West Pakistan
huangpiense: after 黄陂区 (Huángpí District), China
issykkulense: after Иссык-Куль (Issyk-kul’), the Russian name for Ысык-Көл/Isıq-Köl, Kyrgyzstan
japonicum: after the Latin japonicum, meaning “Japanese”
kasokeroense: after Kasokero Cave, Uganda
keterehense: after Ketereh, Malaysia
khani: after غازی خان میرانی (Ghazi Khan), the person after whom ڈيره غازی خان (Dera Ghazi Khan), Pakistan, was named
lusakaense: after Lusaka, Zambia
macquariense: after Macquarie Island, Australia
meramense: after Meram, Turkey
nairobiense: after Nairobi, Kenya
Nairoviridae: from Nairobi sheep disease virus
Norwavirus: from Norway virus
Ocetevirus: from Oceania and Greek τέμνω (témno), meaning to cross
Orthonairovirus: from the Ancient Greek ὀρθός (orthós), meaning “straight, right, proper” and Nairobi, from Nairobi (Kenya) where Nairobi sheep disease virus was first isolated.
parahaemorrhagiae: after the Greek παρά (para), meaning “beside/near/apart from”, and the Latin haemorrhagia, meaning “hemorrhage"
paratemnopterygis: after the host genus name Paratemnopteryx
peruense: after Peru
qalyubense: after قليوب (Qlīūb/Qalyub), Egypt
randallense: after Randall County, USA
Sabavirus: from South Bay virus
sakhalinense: after Сахалиин (Sakhalin), Russia
sanxiaense: after 三峡 (Sānxiá/Three Gorges), China
Shaspivirus: from Shāyáng spider virus 1
soldadoense: after Soldado Rock, Trinidad and Tobago
songlingense: after Sōnglǐng District (松岭区), China
Striwavirus: from Sānxiá water strider virus 1
sulinaense: after Sulina, Northern Dobruja (Dobrogea de Nord), Romania
tachengense: after 塔城 (Tǎchéng), China
thiaforaense: after Thiafora, Senegal
tomdiense: after Tomdi District, Uzbekistan
tunisense: after Tunis, Tunisia
wenzhouense: after 温州市 (Wēnzhōu), China
Xinspivirus: after Xīnzhōu spider virus
xinzhouense: after 新洲区 (Xīnzhōu District), China
yezoense: after Yezo/Ezo/Yeso (蝦夷), the historical name of Hokkaidō (北海道), Japan
yogueense: after Medina Yogue, Senegal
zirkuense: after زركوه (Zrkūh/Zirku Island), United Arab Emirates
Genus and species demarcation criteria
The availability of at least coding-complete sequences of all genome segments is required for nairovirid classification into a species. Demarcation of genera is based upon considerations of their phylogenetic relationships, significant differences in member virus genome architecture, virion antigenicity, and virus ecology (e.g., host range, pathobiology, and transmission patterns). Demarcation of species is achieved by analysis of the pairwise amino-acid sequence identities of virus RdRPs. Similar to the approach used for other bunyavirals, the naturally occurring diversity of nairovirids has been analyzed by the determination of the intraspecies genetic diversity of Crimean Congo hemorrhagic fever (CCHFV), as it represents a well-investigated nairovirid for which numerous complete or coding-complete genome sequences are available. The analysis of pairwise distances among all available CCHFV RdRP amino acid sequences revealed a maximum of 6.8%. Therefore, the species demarcation criterion for nairovirids was set as <93% identity in the amino acid sequence of the RdRP. Hence, nairovirids with <93% RdRP sequence identity represent different species.
Relationships within the family
Phylogenetic relationships across the family have been estimated using maximum likelihood trees generated from complete N, GPC, and L amino-acid sequences (Figure 2 Nairoviridae).
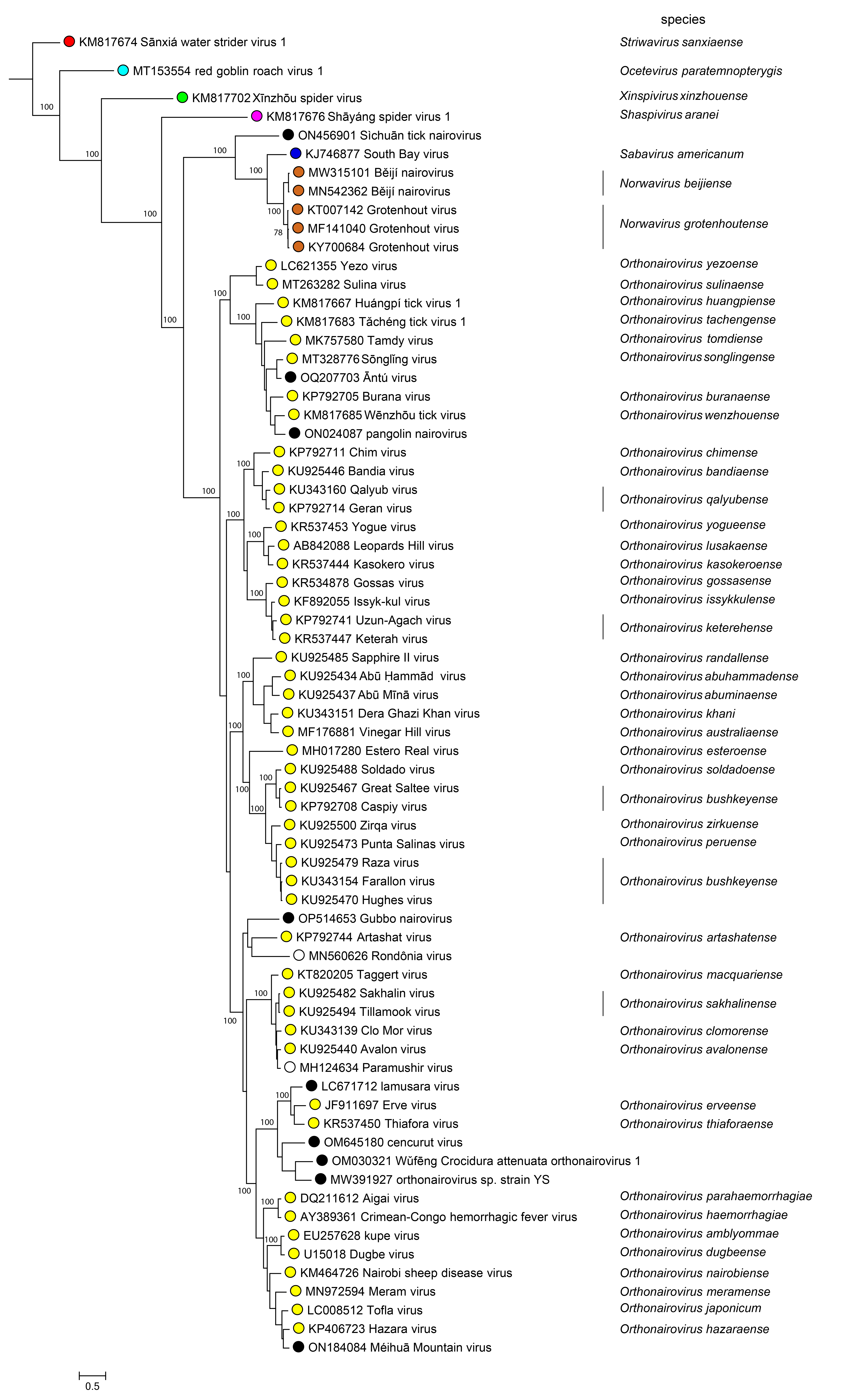 |
| Figure 2 Nairoviridae. The maximum-likelihood phylogenetic tree of nairovirus RdRP amino sequences was inferred using IQtree (Nguyen et al., 2015). Numbers on the nodes represent bootstrap values derived from the ultrafast bootstrap algorithm (Hoang et al., 2018) where these were > 70%. Trees were inferred under the LG+G+I substitution model. Tree branches are proportional to genetic distances between sequences, and the scale bar indicates substitutions per amino acid. |
Relationships with other taxa
Nairovirids are closely related to bunyaviral arenavirids, discovirids, leishbuvirids, mypovirids, phenuivirids, and wupedevirids (Wolf et al., 2018).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Berlin bat nairovirus | L: MN851292* | ||
| Dàlǐ nairo tick virus 1 | L: ON746418 | ||
| Frankliniella occidentalis nairo-like virus 1 | L: MN764155* | (Chiapello et al., 2021) | |
| Fǔshùn Diaea subdola nairovirus 1 | M: MZ210042; L: MZ210041 | ||
| Hángzhōu Paederus fuscipes nairovirus 1 | S: MZ209906; M: MZ209905; L: MZ209904 | ||
| Jí’ān nariovirus | S: ON408084; M: ON408083; L: ON408082 | (Liu et al., 2022) | |
| Línzhī nairo tick virus 1 | L: ON746420 | ||
| Mikuni nairovirus | S: LC795516; L: LC795517 | ||
| Nairoviridae sp. Isolate NH-10 | S: OQ410652; M: OQ410651; L: OQ410650 | ||
| Prackenbach bat nairovirus | L: MN851301* | ||
| Sìchuān tick nairovirus | S: ON456902; L: ON456901 | ScTNV | (Ma et al., 2022) |
| Tōnghuà nairo tick virus 1 | L: ON746414 | ||
| Wittenau bat nairovirus | L: MN851296 | ||
| Xīníng nairo tick virus 1 | L: ON746419 | (Ni et al., 2023) | |
| Yánbiān nairo tick virus 1 | L: ON746415 | ||
| Yīchūn nariovirus | S: ON408109; L: ON408108 | (Liu et al., 2022) | |
| Yúnnán nairo-like virus | S: OR892589; L: OR892588 | YNV | (Wang et al., 2023b) |
| Yúshù nairo tick virus 1 | L: ON746417 |
Virus names and virus abbreviations are not official ICTV designations.
*Coding region sequence incomplete.
Additional unclassified nairovirids that are probable members of established genera are listed under individual genus descriptions.

