Family: Discoviridae
Jens H. Kuhn, Scott Adkins, Katherine Brown, Juan Carlos de la Torre, Michele Digiaro, Holly R. Hughes, Sandra Junglen, Amy J. Lambert, Piet Maes, Marco Marklewitz, Gustavo Palacios, Takahide Sasaya (笹谷孝英), Yong-Zhen Zhang (张永振), and Massimo Turina
*The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Massimo Turina ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2023, updated May 2024
Summary
Discoviridae is a family of negative-sense RNA viruses with genomes of 6.2–9.7 kb (Table 1 Discoviridae). These viruses have been associated with fungi and stramenopiles. The family includes a single genus with six species for seven viruses. The discovirid genome consists of three monocistronic RNA segments with open reading frames (ORFs) that encode a nucleoprotein (NP), a nonstructural (Ns) protein, and a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain.
Table 1 Discoviridae. Characteristics of members of the family Discoviridae
| Characteristic | Description |
| Example | Penicillium discovirus (S: MF142460; M: MF142459; L: MF142458), species Orthodiscovirus missouriense |
| Virion | Unknown |
| Genome | 6.2–9.7 kb of tri-segmented negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Peronosporaceaen stramenopiles, eurotiomycete and sordariomycete fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Hareavirales; the family includes the genus Orthodiscovirus and six species |
Virion
Morphology
Unknown.
Nucleic acid
Discovirids have three segments (small [S], medium [M], and large [L]) of linear negative-sense RNA with a total length of 6.2–9.7 kb (S segment: about 1.1–1.2 kb; M segment: about 1.7–1.9 kb; and L segment: about 3.4–6.5 kb) (Nerva et al., 2019a, Nerva et al., 2019b, Chiapello et al., 2020).
Genome organization and replication
Viruses of the family Discoviridae have a tri-segmented genome that encodes an NP, an Ns protein, and an L protein containing an RNA-directed RNA polymerase (RdRP) domain (Nerva et al., 2019a, Nerva et al., 2019b, Chiapello et al., 2020) (Figure 1 Discoviridae).
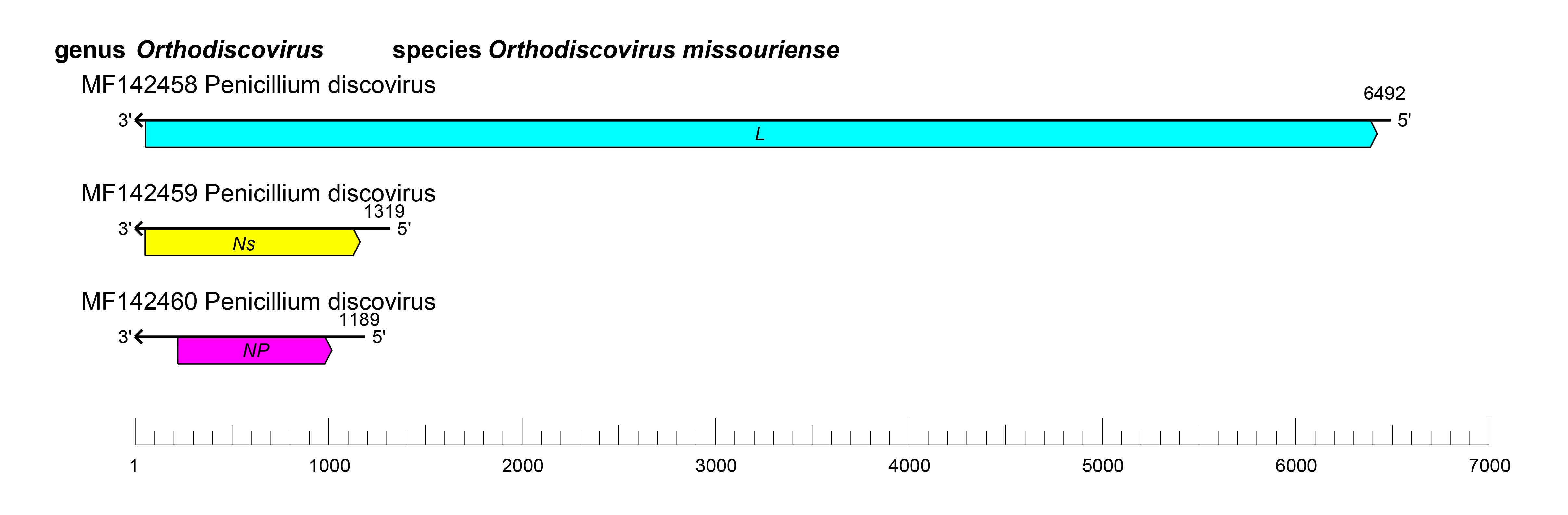 |
| Figure 1 Discoviridae. Genome organization Penicillium discovirus. ORFs are colored according to the predicted protein function (L, large protein gene; NP, nucleoprotein gene; Ns, nonstructural protein gene). |
Biology
The six classified discovirids have been detected in fungi and stramenopiles:
- Coniothyrium diplodiella negative-stranded RNA virus 1 (CdNSRV1) was discovered in sordariomycete fungi (melanconidaceaen Coniella diplodiella (Speg.) Petr. & Syd. (1927)) in Italy (Nerva et al., 2019b)
- Plasmopara viticola lesion associated mycobunyavirales-like virus 9 (PvLAM-LV9) was discovered in oomycetes (peronosporaceaen Plasmopara viticola (Berk. & M.A. Curtis) Berl. & De Toni (1888)) in Italy (Chiapello et al., 2020)
- Plasmopara viticola lesion associated mycobunyavirales-like virus 8 (PvLAM-LV8) was discovered in oomycetes (peronosporaceaen Plasmopara viticola (Berk. & M.A. Curtis) Berl. & De Toni (1888)) in Italy (Chiapello et al., 2020)
- Plasmopara viticola lesion associated mycobunyavirales-like virus 4 (PvLAM-LV4) was discovered in oomycetes (peronosporaceaen Plasmopara viticola (Berk. & M.A. Curtis) Berl. & De Toni (1888)) in Italy (Chiapello et al., 2020)
- Penicillium discovirus (PDV) was discovered in eurotiomycete fungi (trichocomaceaen Penicillium atramentosum Thom, C. 1910) (unpublished)
- Penicillium roseopurpureum negative ssRNA virus 1 (PrNssV1) was discovered in eurotiomycete fungi (trichocomaceaen Penicillium roseopurpureum Dierckx, R.P. 1901) in Italy (Nerva et al., 2019a)
- rice dwarf-associated bunya-like virus has been found in rice (poacean Oryza sativa L.) in China (Wang et al., 2022a)
Unclassified viruses related to discovirids are possibly associated with dothideomycete fungi (pleosporaceaen Alternaria tenuissima Samuel Paul Wiltshire (1933)) in Italy (Wang et al., 2022b), leotiomycete fungi (sclerotiniaceaen Botrytis cinerea Pers. (1794) and erysiphaceaen Erysiphe necator (Schwein.) Burrill)) in Spain (unpublished and (Donaire et al., 2016, Neri et al., 2022, Wang et al., 2022b)), and in sordariomycete fungi (nectriaceaen Fusarium poae (Peck) Wollenw., 1913) (unpublished; (Wang et al., 2022b)). Sequence-read archive analysis indicates that the true diversity of discovirids is vastly underestimated (Neri et al., 2022).
Derivation of names
coniellae: from the host genus Coniella
Discoviridae: from the software DiscVir
hispaniae: from the Latin Hispania, meaning “Spain”
iberiae: from the Latin Hiberia (originating from the Ancient Greek Ἰβηρία [Ibēríā]), meaning “Spain”
missouriense: from Missouri, USA
Orthodiscovirus: from the Greek ortho, meaning “straight” and disco from the software DiscVir
penicillii: from the host genus Penicillium
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Members of different species have RdRP amino acid sequence identities of <80%.
Relationships within the family
Phylogenetic relationships of members of the family Discoviridae are shown in Figure 2 Discoviridae.
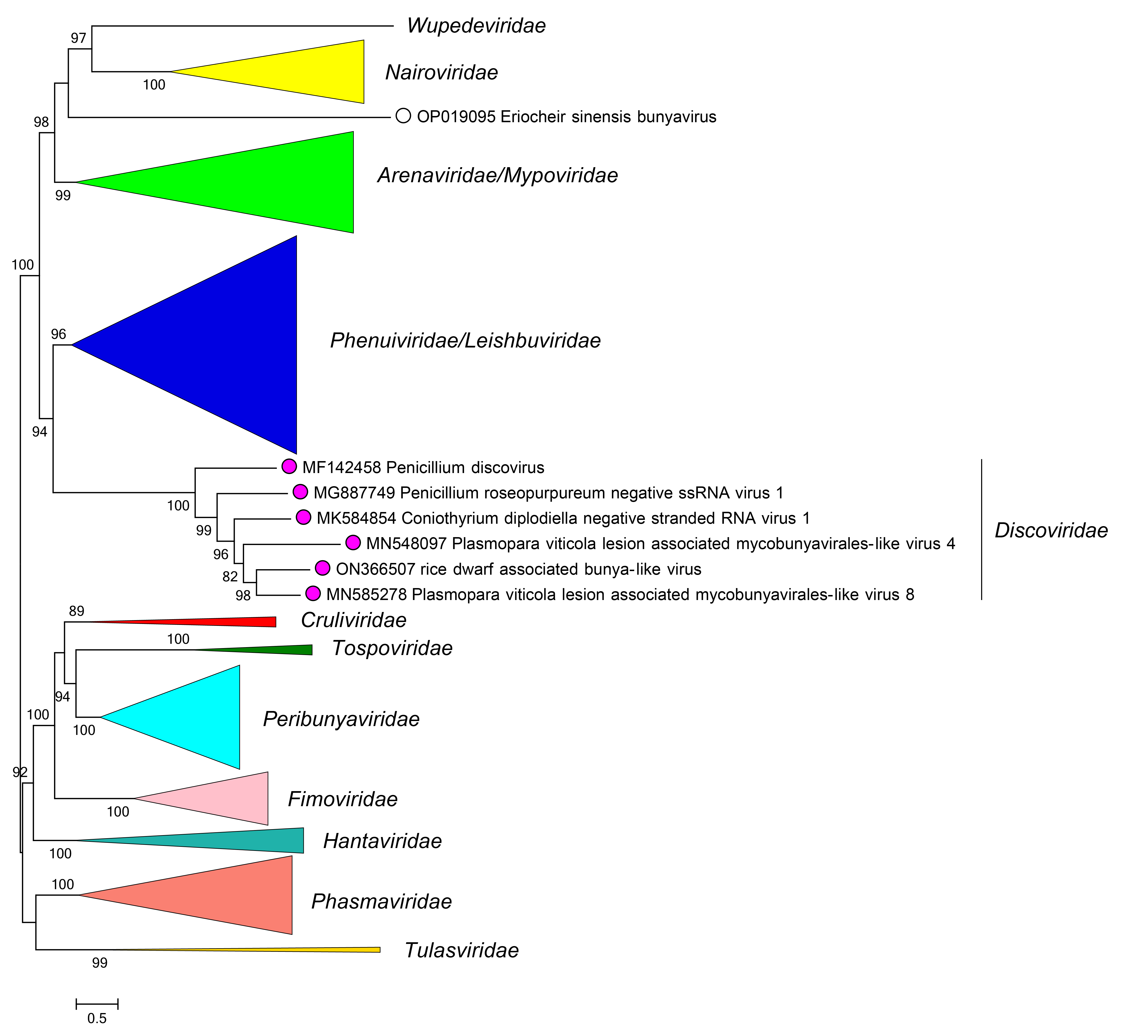 |
| Figure 2 Discoviridae. Phylogenetic relationships of viruses in the family Discoviridae. L protein sequences were aligned using MUSCLE and a maximum likelihood tree was produced using FastTree with default settings. Branches for other families are collapsed. Numbers at nodes indicate bootstrap support where this was > 70%. |
Relationships with other taxa
Viruses in the family Discoviridae are most closely related to bunyaviral arenavirids, leishbuvirids, mypovirids, nairovirids, phenuivirids, and wupedevirids (Huang et al., 2019).

