Family: Closteroviridae
Genus: Crinivirus
Distinguishing features
The genus comprises species whose members are transmitted by whiteflies. Virions are 650–900 nm long and have a bipartite genome, but potato yellow vein virus (PYVV) has a tripartite genome.
Virion
Morphology
See discussion under family description.
Physicochemical and physical properties
See discussion under family description.
Nucleic acid
The virions of members with a bipartite genome contain a single molecule of linear, positive-sense, single-stranded RNA of 7,801 to 9,127 nt (RNA1) and another of 7,903 to 8,530 nt (RNA2). In the case of PYVV, the tripartite genome consists of single-stranded RNAs of 8,035 nt (RNA1), 5,339 nt (RNA2) and 3,892 nt (RNA3).
Proteins
Structural proteins consist of a coat protein (CP) and a minor CP (CPm), with masses ranging from 22 to 29 kDa (CP) and 53 to 80 kDa (CPm), according to virus.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
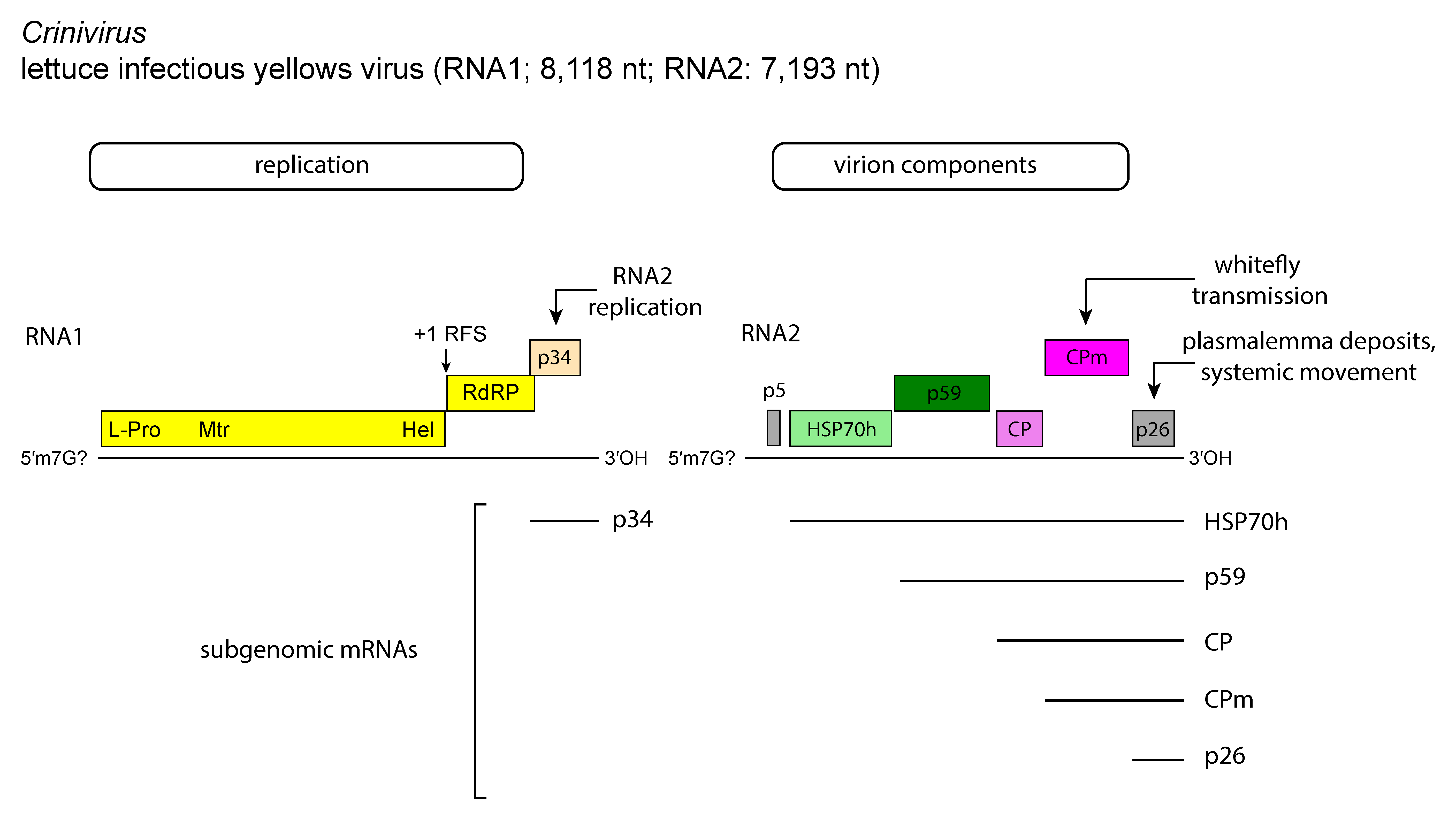
The genome of most criniviruses [e.g. lettuce infectious yellows virus (LIYV)] is divided between two linear, positive-sense, single-stranded RNAs totaling 15.6–17.9 kb (Figure 1. Crinivirus), but PYVV possesses a tripartite genome. All molecules are needed for infectivity and are separately encapsidated. RNA1 of LIYV contains three ORFs, i.e. the ORF1a–ORF1b complex plus a 3′-proximal ORF coding for a 32 kDa protein with no similarity to any protein in databases. This ORF is similar in size and location to ORF2 of citrus tristeza virus (CTV) and beet yellow stunt virus (BYSV) but the respective expression products are not related. RNA1 has 5′- and 3′-UTRs of 97 and 219 nt, respectively. As with other members of the family, the ORF1a–ORF1b complex codes for the replication-related proteins including the RdRP. RNA2 has seven ORFs flanked by a 5′-UTR of 326 nt and a 3′-UTR of 187 nt. RNA2 contains the five-gene module, which, however, differs from that of members of the genus Closterovirus by the insertion of an extra gene (ORF4) upstream of the coat protein (CP) gene. The replication of both genomic RNAs of LIYV is asynchronous, with genomic RNA1 and sgRNAs accumulating before genomic RNA2, and the single-stranded RNA-binding protein p34 encoded by RNA1 acting as a trans enhancer of RNA2 replication (Kiss et al., 2013).
 |
|
Figure 1. Crinivirus. Genome organization of lettuce infectious yellows virus (LIYV), a member of the type species of the genus Crinivirus, showing the relative position of the open reading frames and their expression products: UTR, untranslated region; L-Pro, papain-like protease; Mtr, methyltransferase; Hel, helicase; RdRP, RNA-directed RNA polymerase; HSP70h, heat shock protein 70 homolog; ~60 kDa protein; CP, coat protein; CPm, minor coat protein. |
The genome of PYVV consists of: (i) RNA1 (8,035 nt) with three ORFs, i.e. the ORF1a-ORF1b complex and a 7 kDa hydrophobic protein containing a potential transmembrane helix; (ii) RNA2 (5,339 nt) with five predicted ORFs encoding the HSP70h; a 7 kDa protein similar to a comparable protein of cucurbit yellow stunting disorder virus (CYSDV); the ~60 kDa protein; a 9.8 kDa product with no significant similarity to any other sequence in database; and the 28.2 kDa putative CP; (iii) RNA3 (3,892 nt) with three potential ORFs encoding a 4 kDa protein with no counterpart with other proteins in the family and no significant sequence homology in databases; the 77.5 kDa minor coat protein (CPm), and a 26.4 kDa protein present in other members of the genus. In all criniviruses, the order of the CP and CPm ORFs is similar to that in members of the genera Ampelovirus and Velarivirus but reversed compared to that of members of the genus Closterovirus. Sweet potato chlorotic stunt virus (SPCSV) and tomato chlorosis virus (ToCV) have a particularly large CPm (75–80 kDa) compared to LIYV (53 kDa). Replication occurs in the cytoplasm, likely in association with membranous vesicles derived from the endoplasmic reticulum or from vesiculated mitochondria. Structural and non-structural proteins are similar in type and function to those reported for members of the genus Closterovirus. Both genomic RNAs of ToCV encode RNA silencing suppressors, e.g. the p22 protein in RNA1, and the CP and CPm in RNA2. Suppressor activity is also displayed by the p25 protein of CYSDV, and by the viral RNAse III and the p22 gene present in a few isolates of SPCSV. The LIYV-encoded p26 is involved in systemic plant infection and localizes to plasmodesmata (Qiao et al., 2018).
Biology
Criniviruses infect primarily herbaceous hosts, in which they induce extensive chlorosis to yellow discoloration of the leaves, often accompanied by stunting. They are transmitted semi-persistently by whiteflies of the genera Trialeurodes and Bemisia (Tzanetakis et al., 2013). Persistence and specificity of transmission by their respective vectors have been used as characters for species differentiation. Thus, the viruses of “group 1” [PYVV, blackberry yellow vein-associated virus (BYVaV), beet pseudoyellows virus (BPYV) and strawberry pallidosis-associated virus (SpaV)] are transmitted by T. vaporariorum, viruses of "group 2” [ToCV, SPCSV, CYSDV and bean yellow disorder virus (BYDV)] by B. tabaci, whereas one of the viruses of “group 3” is transmitted by B. tabaci (LIYV) and the other by T. vaporariorum (TICV). These three groups were identified by comparative phylogenetic analyses of RdRP amino acid sequences. None of the viruses is transmitted through seed or mechanically. Geographical distribution varies from restricted (e.g. BYVaV) to very wide. The membranous vesicles with a fibrillar content derive from the endoplasmic reticulum and/or from vesiculated mitochondria (CYSDV).
Antigenicity
Monoclonal antibodies have been produced to proteins of SPCSV. Antisera have been raised from structural and nonstructural proteins produced as fusion proteins in bacterial expression systems (SPCSV and LIYV) or from CPs [tomato infectious chlorosis virus (TICV), LIYV, lettuce chlorosis virus (LCV) and ToCV]. Generally, there are no detectable serological relationships between members of different species. TICV and ToCV, however, are distantly serologically related.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Particle size.
- Size of CP, as determined by deduced amino acid sequence data.
- Serological specificity using discriminatory monoclonal or polyclonal antibodies.
- Genome structure and organization (number and relative location of the ORFs).
- Amino acid sequence of relevant gene products (RdRP, CP, HSP70h) differing by more than 25%.
- Vector species and specificity.
- Magnitude and specificity of natural host range.
- Cytopathological features (i.e., aspect of inclusion bodies and origin of cytoplasmic vesicles).
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
cucurbit chlorotic yellows virus |
AB523788.1, AB523789.1 |
CCYV |
Virus names and virus abbreviations are not official ICTV designations.

