Family: Closteroviridae
Genus: Closterovirus
Distinguishing features
Members of the genus have particle lengths > 1,200 nm and a single-stranded monopartite RNA genome of 14.5–19.3 kb, in which the minor coat protein (CPm) gene is located upstream of the coat protein (CP) gene. Transmission is by aphids.
Virion
Morphology
Particle morphology largely conforms to that of other members of the family, with virions ranging from 1,350 to 2,000 nm in length. Citrus tristeza virus (CTV) has additional smaller than full-length particles that may encapsidate sub-genomic or multiple species of defective RNAs (D-RNAs) containing all of the cis-acting sequences required for replication. Subgenomic RNAs (sgRNAs) may be involved in the construction of recombinant D-RNAs.
Physicochemical and physical properties
According to the species, virus infectivity is inactivated at temperatures between 40 and 55 °C, is retained for 1 to 4 days at room temperature, up to one year in frozen sap, two years in dried leaf material, five years in lyophilized preparations stored at −20 °C, and is destroyed at a pH lower than 6. The A260/A280 ratio is around 1.20 but for some viruses [beet yellows virus (BYV), carnation necrotic fleck virus (CNFV), burdock yellows virus (BuYV)] the virions lack tryptophan, which results in a higher ratio (1.4–1.8). S20w ranges from 130 (BYV) to 140 (CTV), buoyant density is 1.33 g cm−3 in CsCl (BYV and CTV) and 1.257 g cm−3 in Cs2SO4 (CTV).
Nucleic acid
Virions contain a single molecule of linear, positive-sense, single-stranded RNA of 14.5 to 19.3 kb. Multiple dsRNA species occur in infected tissues, the largest of which is usually the replicative form of the entire genome. sgRNAs generate a range of smaller dsRNAs. With CTV, the presence of D-RNA makes the dsRNA pattern of virus isolates more complex than that of other members of the genus.
Proteins
Structural proteins consist of a CP and minor CP (CPm), with a mass ranging from 22 to 25 kDa (CP) and 23 to 27 kDa (CPm), according to virus. The CPm is required for the assembly of the 5′-extremity of the virion, and the 60 kDa protein is required for incorporation of both HSP70h and CPm into virions, which also incorporate a 20 kDa protein that may form the tip segment of the virion head.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Members of the genus Closterovirus whose genomes have been sequenced show three types of genome organization exemplified by those of BYV (Figure 1. Closterovirus), CTV (Figure 2. Closterovirus) and beet yellow stunt virus (BYSV): (i) the BYV genome contains eight ORFs flanked by 5′- and 3′-UTRs of 107 and 181 nt, respectively; (ii) the genome of CTV has 12 ORFs and UTRs of 107 nt at the 5′-end and 275 nt at the 3′-end, and differs from the BYV genome in having two papain-like protease domains in the ORF1a-encoded protein, an extra 5′-proximal ORF (ORF2) encoding a 33 kDa product with no similarity to any other protein in databases, and two extra 3′-proximal ORFs (ORF9 and ORF11); and (iii) the genome of BYSV has 10 ORFs and a 3′-UTR of 241 nt, intermediate between that of the BYSV and CTV UTRs. A further difference with the BYSV genome rests in the presence of an extra ORF (ORF2) encoding a 30 kDa polypeptide with no similarity to any other protein in databases. This ORF is located downstream of ORF1b, i.e. in the same position as the unrelated ORF2 of CTV. Thus, organization of the BYSV genome is intermediate between that of BYV and CTV, suggesting that these three viruses might represent three distinct stages in closterovirus evolution. In addition to CP and CPm, all members of the genus encode proteins such as: (i) a large polypeptide (over 300 kDa) containing the conserved domains of papain-like protease (P-Pro), methyltransferase (Mtr), and helicase (Hel); (ii) an aproximately 50 kDa protein with all the sequence motifs of a viral RNA-directed RNA polymerase (RdRP) of the “alpha-like” supergroup of positive-strand RNA viruses; (iii) a 6 kDa hydrophobic protein with membrane-binding properties; (iv) the homolog of the cellular HSP70; and (v) a 55–64 kDa product, referred to as the ~60 kDa protein. Some of the structural and nonstructural proteins function as suppressors of the plant RNA silencing defense machinery. For instance, the CP, p20 and p23 proteins of CTV have suppressor activity, much the same as the homologs of p21 of BYSV, BYV, and grapevine leafroll-associated virus 2 (GLRaV-2). CTV p23 is a unique protein in the family and has a nucleolar localization (Flores et al., 2013, Ruiz-Ruiz et al., 2019). Silencing suppressors contribute to the accumulation of virus particles and are important determinants of pathogenesis.
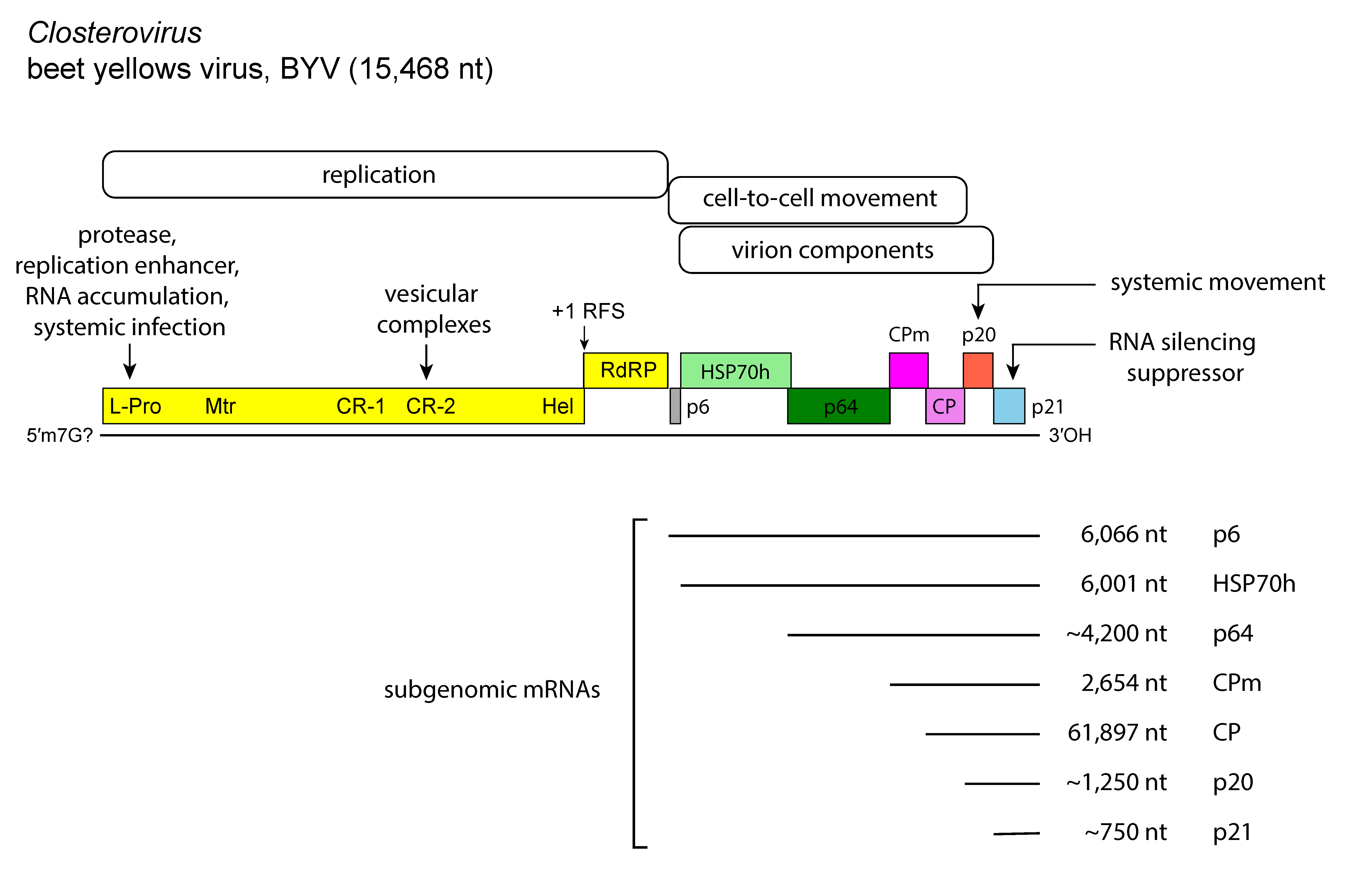 |
|
Figure 1. Closterovirus. Genome organization and strategy of replication of beet yellows virus (BYV) showing the relative position of the open reading frames, their expression products, and the 3′-nested set of sub-genomic RNAs (sgRNAs). L-Pro, leader proteinase (66K); Mtr, methyltransferase (63K); Hel, helicase (100K); RdRP, RNA-directed RNA polymerase; HSP70h, heat shock protein 70 homolog; ~60 kDa protein; CP, coat protein; CPm, minor coat protein. The five boxes under “cell-to-cell movement” represent the five-gene block conserved among most closteroviruses. |
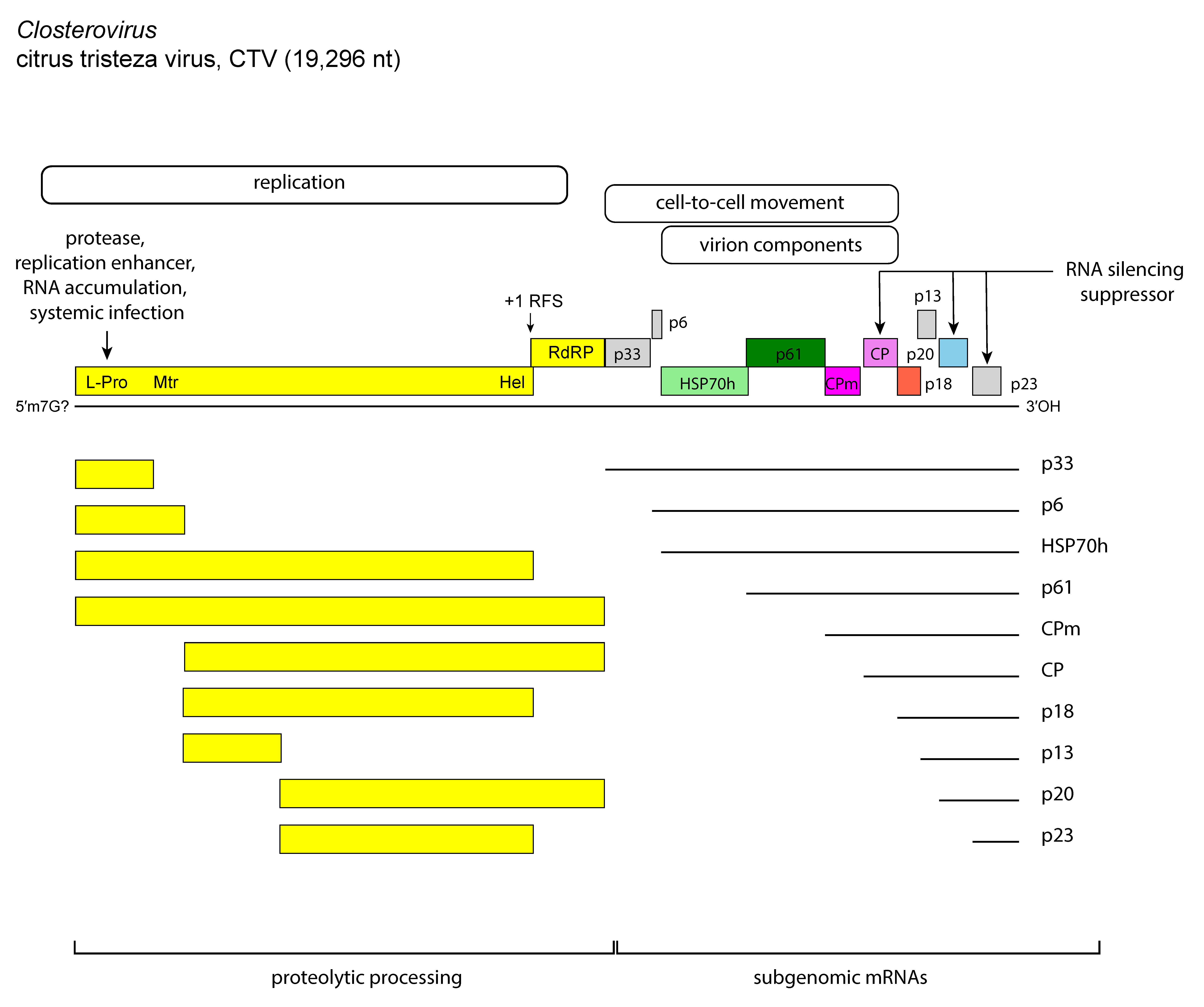 |
|
Figure 2. Closterovirus. Genome organization and expression strategy of citrus tristeza virus (CTV) showing the relative position of the open reading frames and their expression products. The solid lines at the bottom of the figure define the two genomic regions expressed through proteolytic processing of the polyprotein precursor(s) and through the formation of a nested set of 3′-co-terminal sgRNAs. P-Pro, papain-like protease; Mt, methytransferase; Hel, helicase; RdRP, RNA-directed RNA polymerase; HSP70h, heat shock protein 70 homolog; ~60 kDa protein; CPm, minor coat protein; CP, coat protein. |
Biology
Most viruses in the genus infect herbaceous hosts, berry crops and fruit crops. Symptoms are primarily of the discoloration type (yellowing or reddening) or rolling of the leaves. Some CTV strains induce stem pitting. Transmission is by aphids with a semipersistent modality. The range of vectors varies for individual viruses from rather wide to restricted. For instance, BYV is transmitted by 23 aphid species (Myzus persicae and Aphis fabae being the main natural vectors), CTV by seven species (Toxoptera citricida and Aphis gossypii being the most efficient vectors) and a number of other viruses [e.g. CNFV, wheat yellow leaf virus (WYLV), BuYV, mint virus 1 (MV-1)] by a single aphid species, or the vector is unknown (e.g. GLRaV-2). The specific interaction between CTV and its aphid vector Toxoptera citricida indicates the cibarium of the foregut as the virus retention site, and the critical role of a protein-carbohydrate complex for transmission, with CPm, p61 and p65 binding to sugar moieties on the surface of the foregut (Killiny et al., 2016). Members of some species can be transmitted by inoculation of sap, though with difficulty (e.g. CTV, GLRaV-2, BYV), but none are transmitted through seeds. Members of species that infect vegetatively propagated hosts (citrus, grapevine, raspberry) are transmitted by grafting and can be disseminated over long distances with infected propagating material. Geographic distribution ranges from very wide (e.g. CTV, GLRaV-2, BYV) to restricted (e.g. BuYV, MV-1, WYLV).
Antigenicity
No serological relationships are reported among members of different virus species in the genus. Monoclonal antibodies have been produced to BYV, CTV and GLRaV-2 and polyclonal antisera have been raised to BYV, CTV and carrot yellow leaf virus (CYLV) from fusion proteins obtained in bacterial expression systems. Polyclonal antisera have been raised to purified particles of BYV, BYSV, GLRaV-2 and BuYV.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Particle size.
- Size of CP, as determined by deduced amino acid sequence data.
- Serological specificity using discriminatory monoclonal or polyclonal antibodies.
- Genome structure and organization (number and relative location of the ORFs).
- Amino acid sequence of relevant gene products (RdRP, CP, HSP70h) differing by more than 25%.
- Vector species and specificity.
- Magnitude and specificity of natural and experimental host range.
- Cytopathological features (i.e., aspect of inclusion bodies and origin of cytoplasmic vesicles).
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
arracacha virus 1 |
AV1 |
|
|
blackcurrant closterovirus 1 |
BCCV1 |
|
|
burdock yellows virus |
BuYV |
|
|
clover yellows virus |
CYV |
|
|
Dendrobium vein necrosis virus |
DVNV |
|
|
Festuca necrosis virus |
FNV |
|
|
fig leaf mottle-associated virus 1 |
FLMaV1 |
|
|
fig mild mottle-associated virus |
FMMaV |
|
|
Rehmannia virus 1 |
ReV1 |
|
|
wheat yellow leaf virus |
WYLV |
Virus names and virus abbreviations are not official ICTV designations.
*partial sequence

