Family: Chrysoviridae
Ioly Kotta-Loizou, José R. Castón, Robert H.A. Coutts, Bradley I. Hillman, Daohong Jiang, Dae-Hyuk Kim, Hiromitsu Moriyama and Nobuhiro Suzuki
The citation for this ICTV Report chapter is the summary published as Kotta-Loizou et al., (2020):
ICTV Virus Taxonomy Profile: Chrysoviridae, Journal of General Virology, 101:143–144.
Corresponding author: Ioly Kotta-Loizou (i.kotta-loizou13@imperial.ac.uk)
Edited by: Peter Simmonds and Stuart G. Siddell
Posted: January 2018, updated December 2019, August 2021
PDF: ICTV_Chrysoviridae.pdf (2019 version)
This work is dedicated to the memory of our friend and colleague Professor Said Ghabrial, who sequenced the first chrysovirus in 2000 and passed away in November 2018.
Summary
Chrysoviridae is a family of small non-enveloped isometric viruses (approximately 40 nm in diameter) with multi-segmented, double-stranded (ds) RNA genomes (Table 1. Chrysoviridae). Typically, chrysoviruses have four genomic segments, although some have three (collectively known as trichrysoviruses), five (cinquechrysoviruses) or seven (settechrysoviruses) segments. The dsRNA segments are individually encapsidated in separate particles and together comprise 8.9–16.0 kbp of genomic dsRNA. Chrysoviruses infect ascomycetous or basidiomycetous fungi, plants and possibly insects. The two genera, Alphachrysovirus and Betachrysovirus, include 20 and 11 species, respectively. In 2018 the genus Chrysovirus was renamed Alphachrysovirus and a second genus Betachrysovirus was created.
Table 1. Chrysoviridae. Characteristics of members of the family Chrysoviridae.
| Characteristic | Description |
| Example | Penicillium chrysogenum virus ATCC 9480 (dsRNA1: AF296439; dsRNA2: AF296440; dsRNA3: AF296441; dsRNA4: AF296442), species Alphachrysovirus penicillii, genus Alphachrysovirus |
| Virion | Isometric, non-enveloped, approximately 40 nm in diameter |
| Genome | A total of 8.9–16.0 kbp of dsRNA in a multipartite genome (3–7 segments, usually 4) with each segment separately encapsidated |
| Replication | Particles containing both dsRNA and ssRNA can be isolated from infected fungal hosts. Virions accumulate in the cytoplasm |
| Translation | From positive-sense transcripts of genomic dsRNAs |
| Host range | Fungi, plants and possibly insects |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Chrymotiviricetes, order Ghabrivirales: two genera (Alphachrysovirus and Betachrysovirus) including 20 and 11 species, respectively |
Virion
Morphology
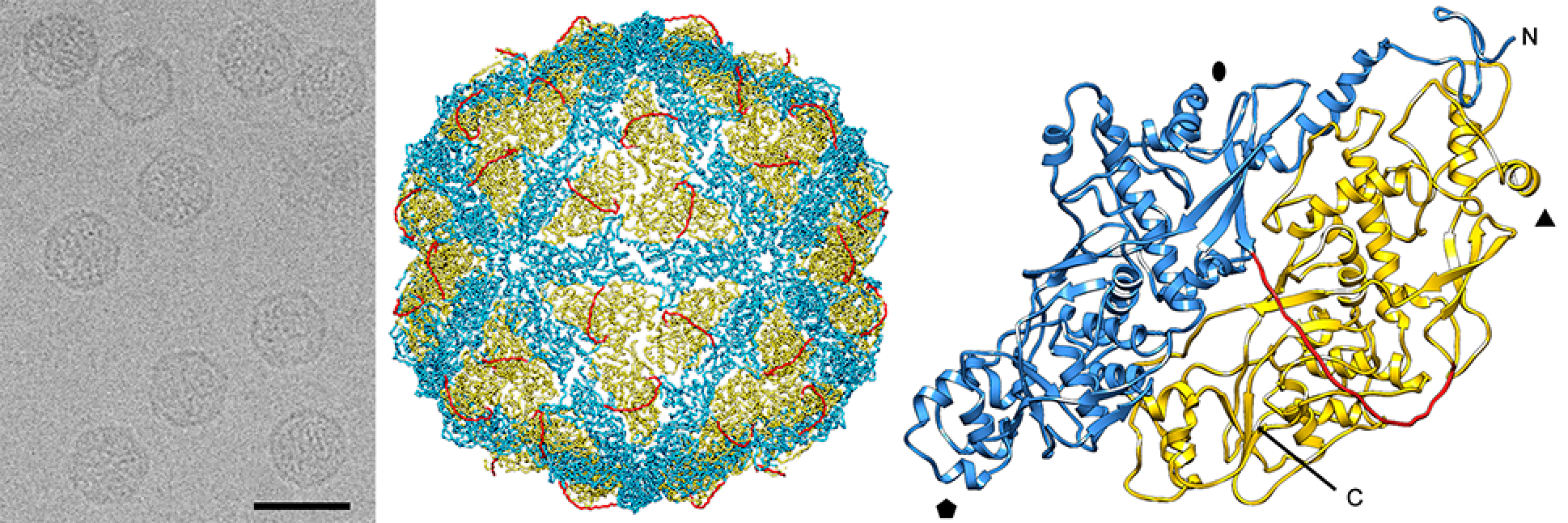
Virions are isometric, non-enveloped, approximately 40 nm in diameter. The capsid of Penicillium chrysogenum virus (PcV) comprises 60 copies of a 109 kDa polypeptide (982 aa) arranged in a T=1 icosahedral lattice (Figure 1. Chrysoviridae). The most prominent features are 12 outward-protruding pentamers. The capsid protein is formed by a repeated α-helical domain, indicative of gene duplication despite a lack of sequence similarity between the two halves (Luque et al., 2010). This domain has a fold that is conserved among dsRNA viruses (Luque et al., 2014). The capsid inner surface has highly positively charged triskelion-shaped areas that maintain the encapsidated genome in close contact with the inner capsid surface. Arrangement of dsRNA might facilitate genome mobility within the capsid for its transcription and replication. It may also have a structural role in capsid stability (Castón et al., 2013).
Physicochemical and physical properties
Virion buoyant density in CsCl is in the range of 1.34–1.39 g cm−3and S20, w is 145–150S (Buck and Girvan 1977, Sanderlin and Ghabrial 1978, Wood and Bozarth 1972).
Nucleic acid
Virions contain from three to seven linear, separately encapsidated, dsRNA segments, ranging in size from 0.8 to 3.7 kbp.
Proteins
The capsids are made up of a single major polypeptide species, the capsid protein (CP; 80–126 kDa). Virion-associated RNA-directed RNA polymerase (RdRP) activity is present.
Genome organization and replication
Chrysoviruses have multipartite genomes comprising three to seven linear dsRNA segments. Each segment is monocistronic; usually dsRNA1 codes for the RNA-directed RNA polymerase (RdRP; P1) and dsRNA2 codes for the major capsid protein (CP; P2). The rest of the virus-encoded proteins are of unknown function. Assignment of numbers 1–4 to PcV and Botryosphaeria dothidea chrysovirus 1 (BdCV1) dsRNAs was made according to their decreasing size. There is, however, some variation in the relative sizes of segments 3 and 4 among other chysoviruses. For example, in the genus Alphachrysovirus, the dsRNA segment 3 of Amasya cherry disease associated chrysovirus (ACDACV) (Covelli et al., 2004), Cryphonectria nitschkei chrysovirus 1 (CnCV1) (Kim et al., 2010), Helminthosporium victoriae virus 145S (HvV145S) (Ghabrial et al., 2013) and Macrophomina phaseolina chrysovirus 1 (MpCV1) (Marzano et al., 2016) is in each case shorter than the corresponding segment 4. Sequence comparisons confirmed that dsRNA3 of ACDACV, CnCV1, HvV145S and MpCV1 are in fact the counterparts of PcV dsRNA4 rather than dsRNA3. Likewise, the dsRNA4 of these three chrysoviruses is the counterpart of PcV dsRNA3. Since PcV was the first chrysovirus to be characterized at the molecular level and to avoid confusion, the protein designations P3 and P4 as used for PcV are adopted and referred to as alphachryso-P3 and alphachryso-P4. Similarly, for members of the genus Betachrysovirus, the protein designations P3 and P4 as used for BdCV1 are adopted and referred to as betachryso-P3 and betachryso-P4.
The virion-associated RdRP catalyzes in vitro end-to-end conservative transcription of each dsRNA to produce mRNA. Virions accumulate in the cytoplasm.
The majority of 5′- and 3′-NCRs are relatively long, up to 698 nt and 865 nt respectively. The sequences of the 5′- and 3′-NCRs are highly conserved among the four-dsRNA segments (Figure 2. Chrysoviridae). In addition to the absolutely conserved 5′- and 3′-termini, a 40–75 bp region with high sequence identity is present in the 5′-NCR of all four PcV dsRNAs (Box1; Figure 2. Chrysoviridae). A second region of strong sequence similarity is present immediately downstream from Box 1 and consists of 30–50 bp containing a reiteration of the sequence “CAA”; it is found to a lesser extent in the 5′-UTRs of BdCV1. The (CAA)n repeats are similar to the enhancer elements present at the 5′-UTRs of tobamoviruses (Gallie and Walbot 1992).
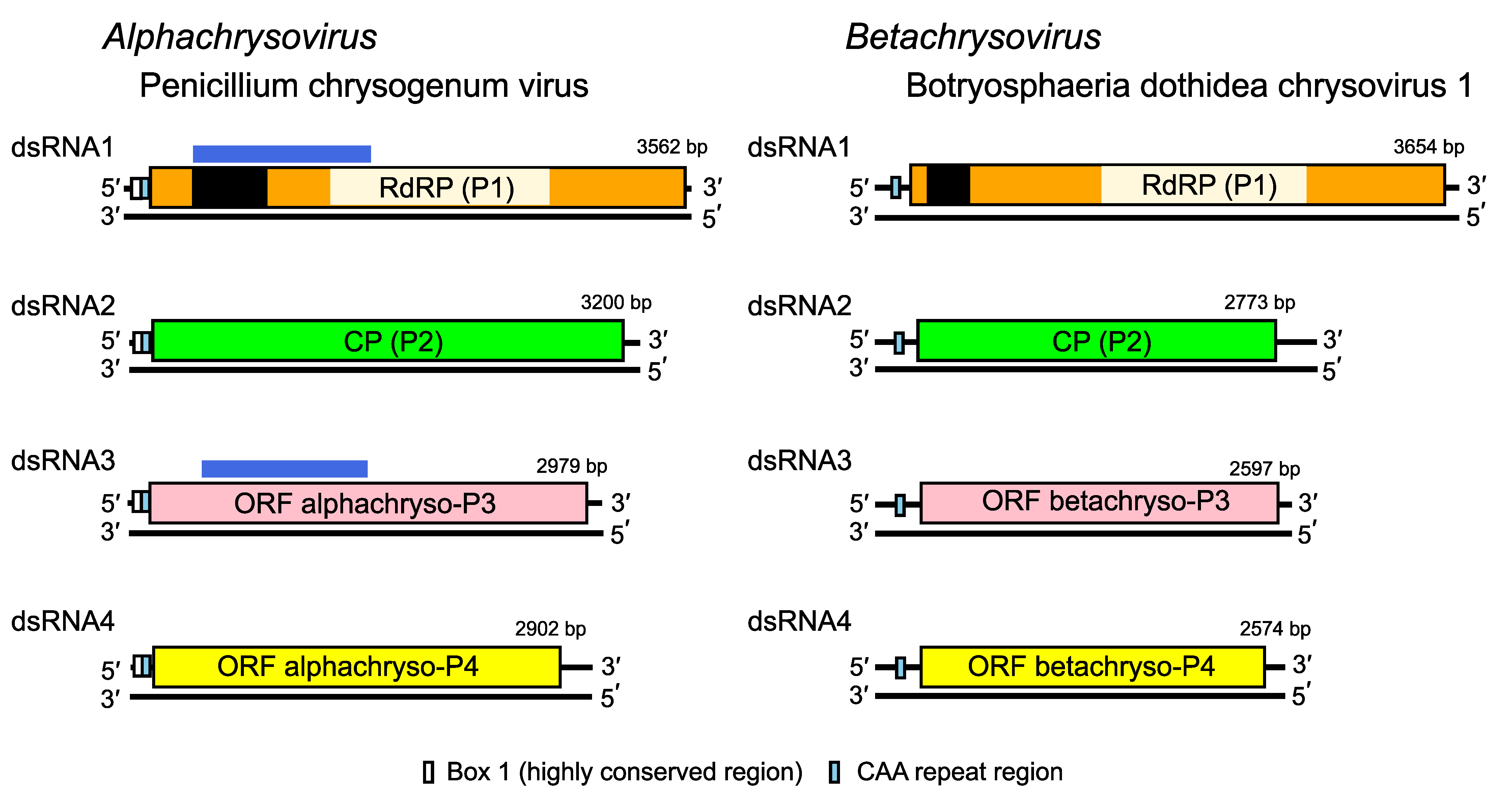 |
| Figure 2. . Chrysoviridae. Genome organization of Penicillium chrysogenum virus (PcV) and Botryosphaeria dothidea chrysovirus 1 (BdCV1). The genomes consist of four dsRNA segments; each is monocistronic. The proteins encoded are represented by rectangular boxes. The light coloured box in dsRNA 1 represents the RdRP_4 motif (PF02123); the dark coloured box in dsRNA 1 represents an independent P-loop NTPase domain. Although the functions of P3 and P4 are unknown (see below) for both viruses, the N-terminal region of alphachryso-P3 shares high sequence similarity with the corresponding N-terminal region of the RdRP (P7/P-loop domain; possibly a nucleotide triphosphate hydrase [NTPase] domain), as indicated by the blue boxes above the ORFs. Alphachryso-P4 is a putative cysteine protease. |
Biology
Chrysoviruses lack an extracellular phase to their life cycle; they are transmitted intracellularly during cell division and sporogenesis (vertical transmission) and by cell fusion following hyphal anastomosis between compatible fungal strains (horizontal transmission) (Ghabrial 2008). Trichrysoviruses (those with three dsRNA segments) mostly infect plants with evidence for vertical transmission through seeds (Zhang et al., 2017, Li et al., 2013). Chrysoviruses are typically associated with latent infections, although some cinquechrysoviruses (five dsRNA segments) cause deleterious effects in their fungal hosts (Urayama et al., 2010, Urayama et al., 2012, Urayama et al., 2014). The definitive host is not known for chrysoviruses identified by next generation sequencing of insect material.
Antigenicity
High affinity antisera are produced in rabbits immunized with purified virions.
Derivation of names
Chryso: chrysós meaning “gold” in Greek, derived from the specific epithet of Penicillium chrysogenum, the fungal host of the prototype strain of the type species, which often produces a golden yellow pigment.
Alpha: the first letter of the Greek alphabet, indicating the first genus of the family Chrysoviridae.
Beta: the second letter of the Greek alphabet, indicating the second genus of the family Chrysoviridae.
Genus demarcation criteria
Genus demarcation criteria:
- phylogenetic analysis
- nucleotide and deduced amino acid sequence data (e.g. with the exception of the RdRP, the viral proteins are non-homologous across genera)
- number of dsRNA genome segments
Relationships within the family
Phylogenetic analysis based on the complete deduced amino acid sequences of the RdRP of members of family Chrysoviridae and related viruses (Figure 3. Chrysoviridae) leads to the identification of two distinct clusters: the first one corresponds to members of the genus Alphachrysovirus and includes viruses with 3 or 4 genomic segments, while the second one comprises chrysoviruses with 4, 5 or 7 genomic segments belonging to the genus Betachrysovirus.
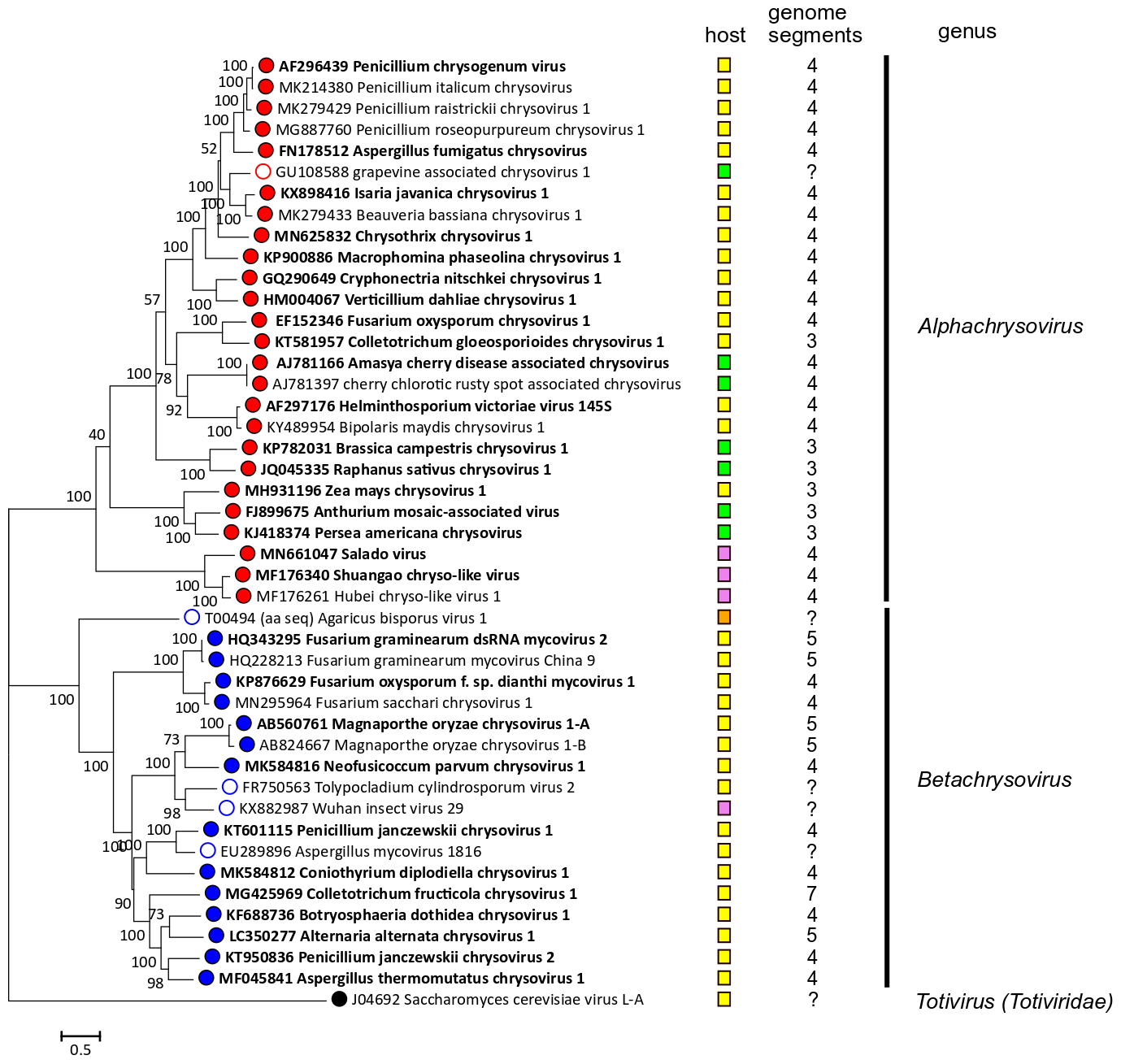 |
| Figure 3. Chrysoviridae. Phylogenetic analysis of members of the family Chrysoviridae and related viruses based on amino acid sequences of their RdRP. A multiple alignment of RdRP amino acid sequences was produced using MUSCLE (Edgar 2004), all positions with less than 30% site coverage were eliminated and the LG+G+I+F substitution model was used and a maximum likelihood phylogenetic tree was created by the program MEGA 6.0 (Tamura et al., 2013). Bootstrap percentages (100 replicates) are shown. Tips labelled with open circles indicate unclassified viruses. Exemplar isolates are indicated by bold text. Virus hosts or associations are indicated by squares coloured yellow (fungi, ascomycetes), orange (fungi, basidiomycetes) green (plants) and pink (invertebrate samples). The number of dsRNA genomic segments is indicated, if known. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Other fungal viruses that possess four dsRNA genome segments include members of the families Quadriviridae (Lin et al., 2012), Polymycoviridae (Kanhayuwa et al., 2015, Kotta-Loizou and Coutts 2017) and the proposed family Alternaviridae (Aoki et al., 2009, Kozlakidis et al., 2013). However, of these only members of the family Quadriviridae show any close phylogenetic relatedness to chrysoviruses in the RdRP gene, as also do members of the families Botybirnaviridae, Megabirnaviridae and Totiviridae. These latter viruses, however, have quite different genome organisations (2, 2 and 1 genome segments respectively). Virion structures are also distinct; the latter possess T=1 capsids made up from 120-subunits of homodimeric or heterodimeric CPs (Luque et al., 2016) instead of 60 copies of a single, internally duplicated CP arranged on a T=1 icosahedral lattice (Figure 1. Chrysoviridae) (Castón et al., 2003).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| grapevine associated chrysovirus 1 | GU108588 | GACV1 |
| Aspergillus mycovirus 1816 | EU289896 | AMV1816 |
| Tolypocladium cylindrosporum virus 2 | FR750563 | TcV2 |
| Agaricus bisporus virus 1 | X94361 | AbV1 |
| Wuhan insect virus 29 | KX882987 | WuIV29 |
Virus names and virus abbreviations are not official ICTV designations.
These viruses have partially sequenced genomes; grapevine associated chrysovirus 1 is related to alphachrysoviruses, while the other viruses are related to betachrysoviruses (Figure 3. Chrysoviridae). The number of segments for each virus is unknown.