References: Myriaviridae
References: Myriaviridae
Di Paola, N., Dheilly, N. M., Junglen, S., Paraskevopoulou, S., Postler, T. S., Shi, M. & Kuhn, J. H. (2022). Jingchuvirales: a new taxonomical framework for a rapidly expanding order of unusual monjiviricete viruses broadly distributed among arthropod subphyla. Appl Environ Microbiol 88, e0195421. [PubMed]
Citation: Myriaviridae
Citation: Myriaviridae
A summary of this ICTV Report chapter has been published as an ICTV Virus Taxonomy Profile article in the Journal of General Virology, and should be cited when referencing this online chapter as follows:
Authors: Myriaviridae
Authors: Myriaviridae
Jens H. Kuhn
Integrated Research Facility at Fort Detrick
National Institute of Allergy and Infectious Diseases
National Institutes of Health
Fort Detrick
Frederick
Maryland
USA
Email: [email protected]
Nolwenn M. Dheilly
Pathogen Discovery Laboratory
Institut Pasteur
Université Paris Cité
75015 Paris
France
Genus: Myriavirus
Family: Myriaviridae
Genus: Myriavirus
Distinguishing features
There is only one genus in the family and so the distinguishing features correspond the family description.
Species demarcation criteria
Not applicable (this only genus includes one species).
Family: Myriaviridae
Family: Myriaviridae
Jens H. Kuhn, Nolwenn M. Dheilly, Sandra Junglen, Sofia Paraskevopoulou (Σοφία Παρασκευοπούλου), Mang Shi (施莽), and Nicholas Di Paola
The citation for this ICTV Report chapter is the summary published as:
Myriaviridae
Myriaviridae Table of Contents
Species List: Crepuscuviridae
Species List: Crepuscuviridae
Virus names, the choice of exemplar isolates, and virus abbreviations, are not official ICTV designations
Exemplar isolate of the species
Resources: Crepuscuviridae
Resources: Crepuscuviridae
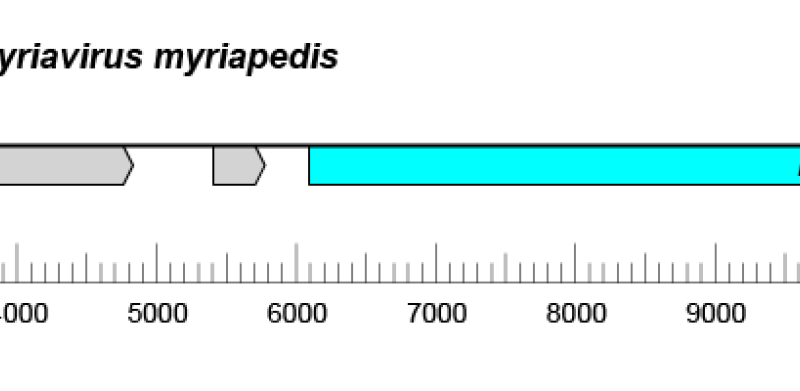
Figure 2.Crepuscuviridae
References: Crepuscuviridae
References: Crepuscuviridae
Di Paola, N., Dheilly, N. M., Junglen, S., Paraskevopoulou, S., Postler, T. S., Shi, M. & Kuhn, J. H. (2022). Jingchuvirales: a new taxonomical framework for a rapidly expanding order of unusual monjiviricete viruses broadly distributed among arthropod subphyla. Appl Environ Microbiol 88, e0195421. [PubMed]