Family: Poxviridae
Colin J. McInnes, Inger K. Damon, Geoffrey L. Smith, Grant McFadden, Stuart N. Isaacs, Rachel L. Roper, David H. Evans, Clarissa R. Damaso, Olivia Carulei, Lyn M. Wise and Elliot Lefkowitz
The citation for this ICTV Report chapter is the summary published as McInnes et al., (2023):
McInnes CJ, Damon IK, Smith GL, McFadden G, Isaacs SN, Roper RL, Evans DH, Damaso CR, Carulei O, Wise LM, Lefkowitz EJ. ICTV Virus Taxonomy Profile: Poxviridae 2023. J Gen Virol. 2023 May;104(5). doi: 10.1099/jgv.0.001849. PMID: 37195882.
Corresponding author: Colin McInnes ([email protected])
Edited by: Arvind Varsani and Stuart Siddell
Posted: April 2023
Summary
Poxviridae is a family of enveloped, generally oval or brick-shaped viruses, 220–450 nm long, 140–260 nm wide and 140–260 nm thick with a genome comprising a single linear molecule of dsDNA of 128–375 kbp with covalently-closed ends (Table 1.Poxviridae). The family includes two subfamilies. Members of the subfamily Chordopoxvirinae infect vertebrates and are classified in the genera Avipoxvirus, Capripoxvirus, Centapoxvirus, Cervidpoxvirus, Crocodylidpoxvirus, Leporipoxvirus, Macropopoxvirus, Molluscipoxvirus, Mustelpoxvirus, Orthopoxvirus, Oryzopoxvirus, Parapoxvirus, Pteropopoxvirus, Salmonpoxvirus, Sciuripoxvirus, Suipoxvirus, Vespertilionpoxvirus and Yatapoxvirus. Members of the Chordopoxvirinae include important pathogens of humans (variola virus, the causative agent of smallpox), livestock animals and wildlife, including aquatic species. Infections typically result in the formation of lesions, skin nodules, or disseminated rash and can be fatal in some cases. Members of the subfamily Entomopoxvirinae infect insects, and are classified into four genera; Alphaentemopoxvirus, Betaentemopoxvirus, Deltaentomopoxvirus and Gammaentemopoxvirus. Generally, members of the above entompoxvirus genera infect a different order of insect, respectively Coleoptera, Lepidoptera, Orthoptera and Diptera. Infection with members of the subfamily Entompoxvirinae is invariably fatal, but disease progression is generally slow. The subfamily Entomopoxvirinae includes one species, Diachasmimorpha entomopoxvirus that is unassigned to a genus.
Table 1.Poxviridae Characteristics of members of the family Poxviridae
| Characteristic | Description |
| Example | vaccinia virus (AY243312), species Orthopoxvirus vaccinia |
| Virion | Generally oval or brick-shaped, 220–450 nm × 140–260 nm × 140–260 nm with a lipoprotein surface membrane displaying tubular or globular units or a regular spiral filament. |
| Genome | Single linear molecule of dsDNA with covalently-closed ends. 128–375 kbp. |
| Replication | Cytoplasmic, mediated by virus-encoded proteins with self priming. DNA replicated as long concatemers that are resolved by a viral HJ endonuclease. |
| Translation | Polyadenylated, capped mRNA transcripts, synthesized from both DNA strands by virus-encoded RNA polymerase. |
| Host range | Vertebrates and arthropods. |
| Taxonomy | Realm Varidnaviria, kingdom Bamfordvirae, phylum Nucleocytoviricota, class Pokkesviricetes, order Chitovirales: 2 subfamilies, 22 genera and 56 species |
Virion
Morphology
Virions are somewhat pleomorphic, generally brick-shaped (220–450 nm long×140–260 nm wide×140–260 nm thick) with a lipoprotein surface membrane displaying tubular or globular units (10–40 nm). Virions can also be ovoid (250–300 nm long×160–190 nm diameter) with a surface membrane possessing a regular spiral filament (10–20 nm in diameter) (Figure 1.Poxviridae). Negatively-stained electron microscopy images show that the surface membrane encloses a biconcave or cylindrical core that contains the genome DNA and proteins organized in a nucleoprotein complex. One or two lateral bodies appear to be present in the concave region between the core wall and a membrane (Condit et al., 2006). One model suggests that the nucleoprotein complex might be cylindrical, folded at least twice along the long virion axis to form a Z-structure, which presents as three circles, arranged linearly when viewed as a section across the short axis. This virion form is known as the mature virion (MV; also known as intracellular mature virus, IMV). Some MV is wrapped by an additional double layer of intracellular membranes (derived from the trans-Golgi or endosomes) to form wrapped virions (WV; also known as intracellular enveloped virus, IEV). WV can be externalized, losing the outermost of the additional membrane layers via fusion with the cell membrane, to form extracellular virions (EV). EV are antigenically distinct from MV, due to the presence of envelope-specific proteins. They can be bound to the cell surface (in a form specifically known as cell-associated enveloped virus, CEV) or released into the extracellular medium (in a form specifically known as extracellular enveloped virus, EEV). While this pathway commonly predominates in the mammalian poxviruses, the avian viruses (e.g. canarypox virus, fowlpox virus and pigeonpox virus) appear to form EV directly by budding of MV through the cell membrane, rather than by intermediate formation of WV. The avian viruses and some mammalian viruses (e.g. cowpox virus, ectromelia virus and raccoonpox virus, but not vaccinia virus or variola virus, and not all isolates) may also sequester MV within inclusion bodies. Other poxviruses (e.g. entomopoxviruses) may be occluded into a preformed inclusion body.
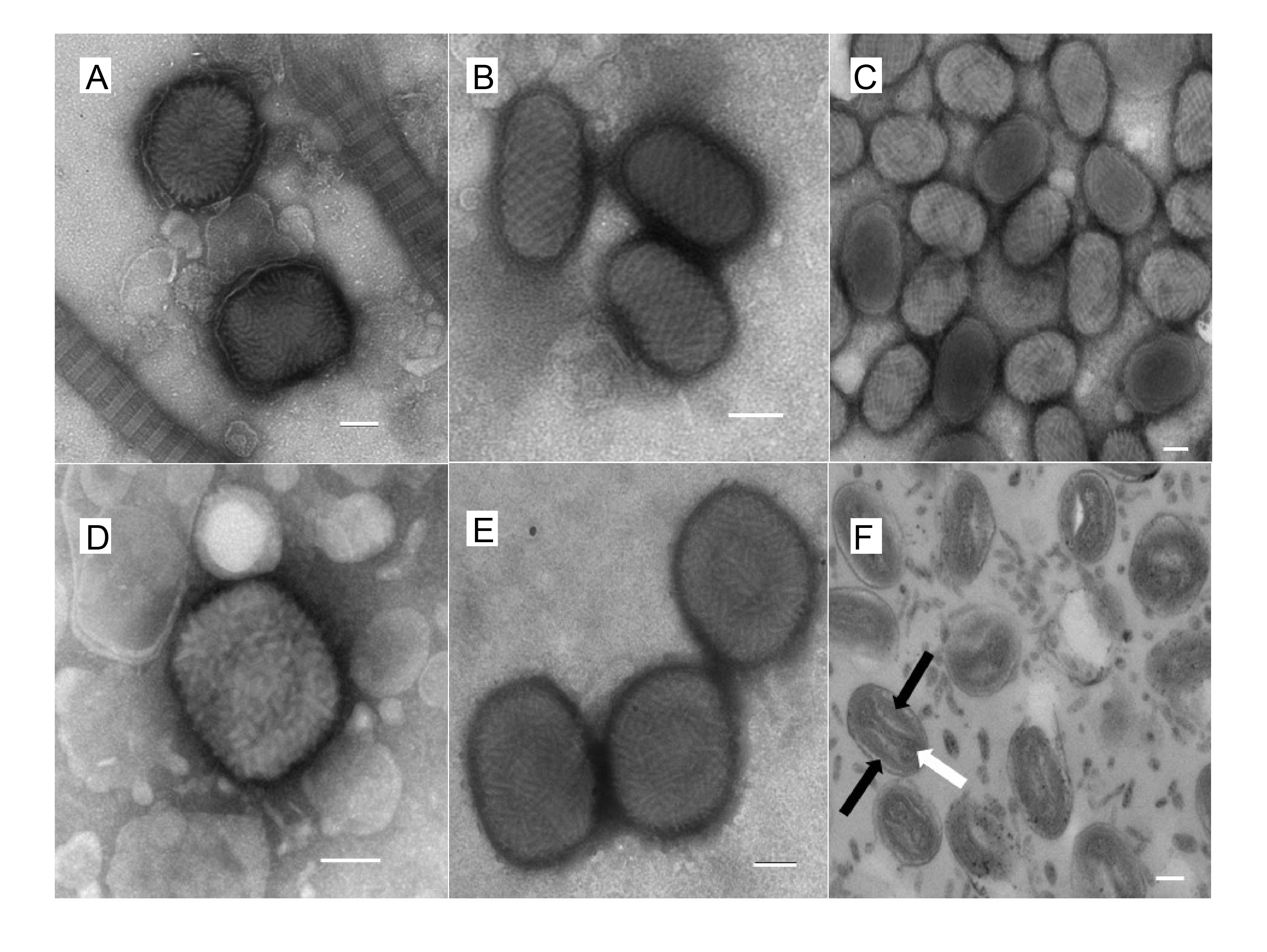 |
| Figure 1.Poxviridae. Morphology of poxviruses visualized with electron microscopy. Electron micrographs of negatively-stained preparations of: (A) myxoma virus (genus Leporipoxvirus) mature virions (the virions are seen alongside collagen fibres); (B) orf virus (genus Parapoxvirus) mature virions; (C) squirrelpox virus mature virions; (D) an orthopoxvirus virion; (E) avipoxvirus virions; and (F) avipoxvirus virions demonstrating the two lateral bodies (black arrows) either side of the “dumbbell” core (white arrow). The bar in each case represents 100 nm. Images courtesy of David J Everest ([email protected]), Animal and Plant Health Agency (APHA), Surrey, UK. |
Physicochemical and physical properties
Particle Mr is 3 ×109; S20,W is about 5000S. Buoyant density of virions is subject to osmotic influences: in dilute buffers it is about 1.16 g cm−3, in sucrose about 1.25 g cm−3, and in CsCl and potassium tartrate about 1.30 g cm−3. Virions tend to aggregate in high salt solution and at high- particle concentrations. Infectivity of some members is resistant to trypsin, but in some cases it is enhanced by trypsin treatment. Some members are insensitive to ether. Generally, virion infectivity is sensitive to common detergents, formaldehyde, oxidizing agents and temperatures greater than 40 °C. The virion surface membrane is removed by nonionic detergents and sulfhydryl reducing reagents. Virions are relatively stable in dry conditions at room temperature; they can be lyophilized with little loss of infectivity.
Nucleic acid
Nucleic acids constitute about 3% of the particle weight. The genome is a single, linear molecule of dsDNA (128–375 kbp) with covalently-closed termini.
Proteins
Poxvirus proteins constitute about 90% of the particle weight. Poxvirus genomes encode 120–300 proteins depending on the virus; about 100 proteins are present in virions. Virus particles contain many enzymes involved in DNA transcription or modification of proteins or nucleic acids. Enveloped virions have virus-encoded polypeptides in the lipid bilayer, which surrounds the particle. Entomopoxviruses may be occluded by a virus-encoded, major structural protein, spheroidin. Similarly, chordopoxviruses may be within inclusion bodies again consisting of a single protein (the A-type inclusion ATI protein family).
Lipids
Lipids constitute about 4% of the poxvirus particle weight. Enveloped poxvirions contain lipids, including glycolipids, which may be modified cellular lipids.
Carbohydrates
Carbohydrates constitute about 3% of the poxvirus particle weight. Certain viral proteins in EV, e.g. haemagglutinin in the envelope of orthopoxviruses, have N- and C-linked glycans.
Genome organization and replication
The poxvirus genome comprises a linear molecule of dsDNA with covalently closed termini; terminal hairpins constitute two isomeric, imperfectly paired, “flip-flop” DNA forms consisting of inverted complementary sequences (Figure 2.Poxviridae). Variably sized, tandem repeat sequence arrays may or may not be present near the ends. In general, conserved proteins essential to virus replication in culture (polymerases and other enzymes, and structural proteins) are encoded in the central region of the genome whereas less conserved, non-essential proteins involved in virus–host responses (immunomodulators, anti-apoptotic proteins, etc.) are encoded in the terminal regions of the genome. Several large protein families are encoded by poxvirus genomes, in some cases with many members. For instance, canarypox virus encodes 51 homologous proteins of the ankyrin repeat family.
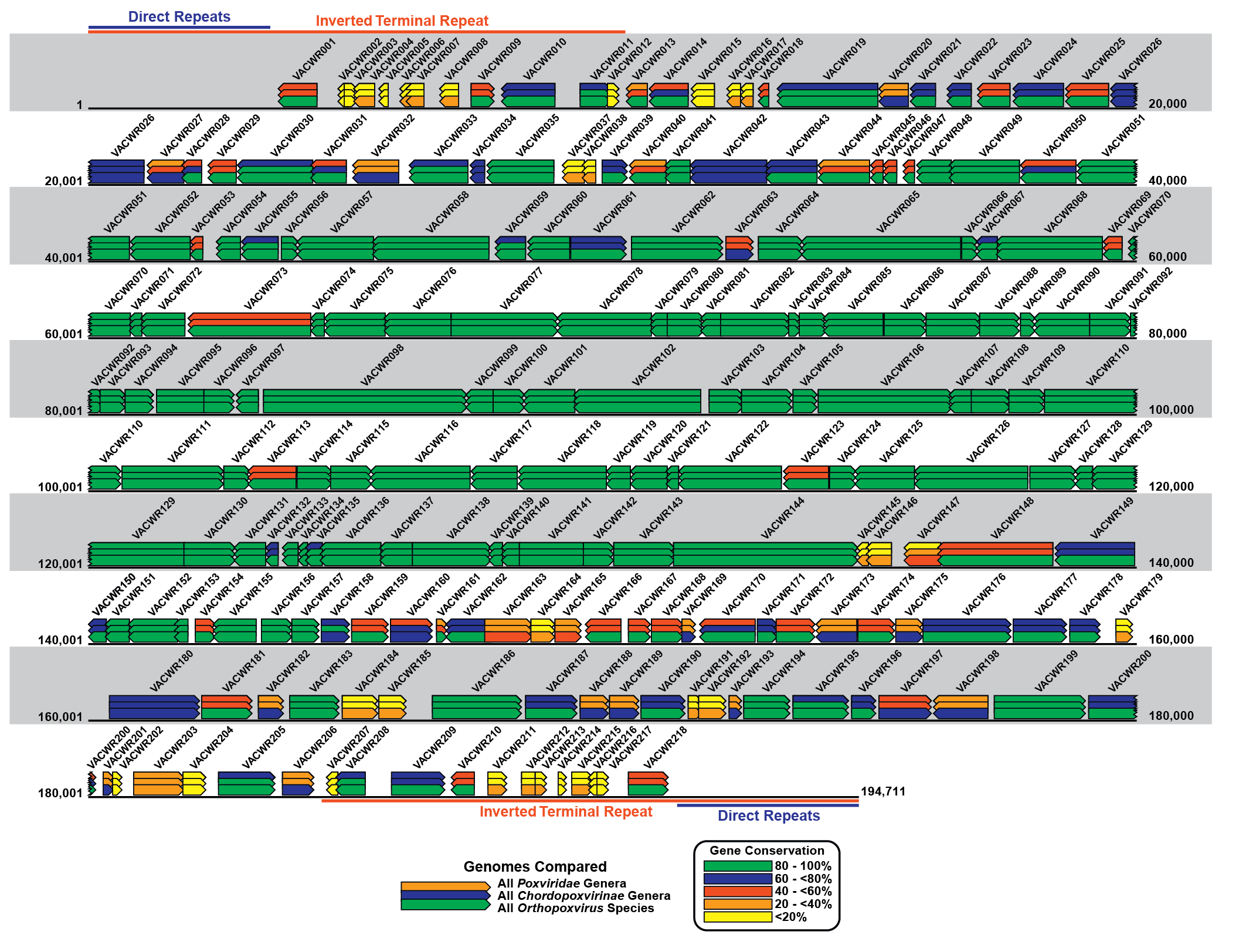 |
| Figure 2.Poxviridae Schematic representation of the poxvirus genome. Schematic representation of the genome of the WR strain of vaccinia virus (VACV-WR: AY243312). The genome is a linear double-stranded molecule with terminal hairpins, inverted terminal repeats (ITR) and a series of direct repeats within the ITRs. Coloured rectangles represent each WR gene and the arrow at one end of each rectangle indicates the direction of transcription for that gene from the DNA template. The VACWR name represents the GenBank locus name for each gene. The coloured overlapping gene rectangles indicate the extent to which each gene is conserved (present or absent) in all poxviruses, vertebrate poxviruses (chordopoxviruses) and orthopoxviruses. The bars are colour-coded according to the percentage of gene conservation across the indicated taxa. |
Replication takes place predominately, if not exclusively, within the cytoplasm (Figure 3.Poxviridae).
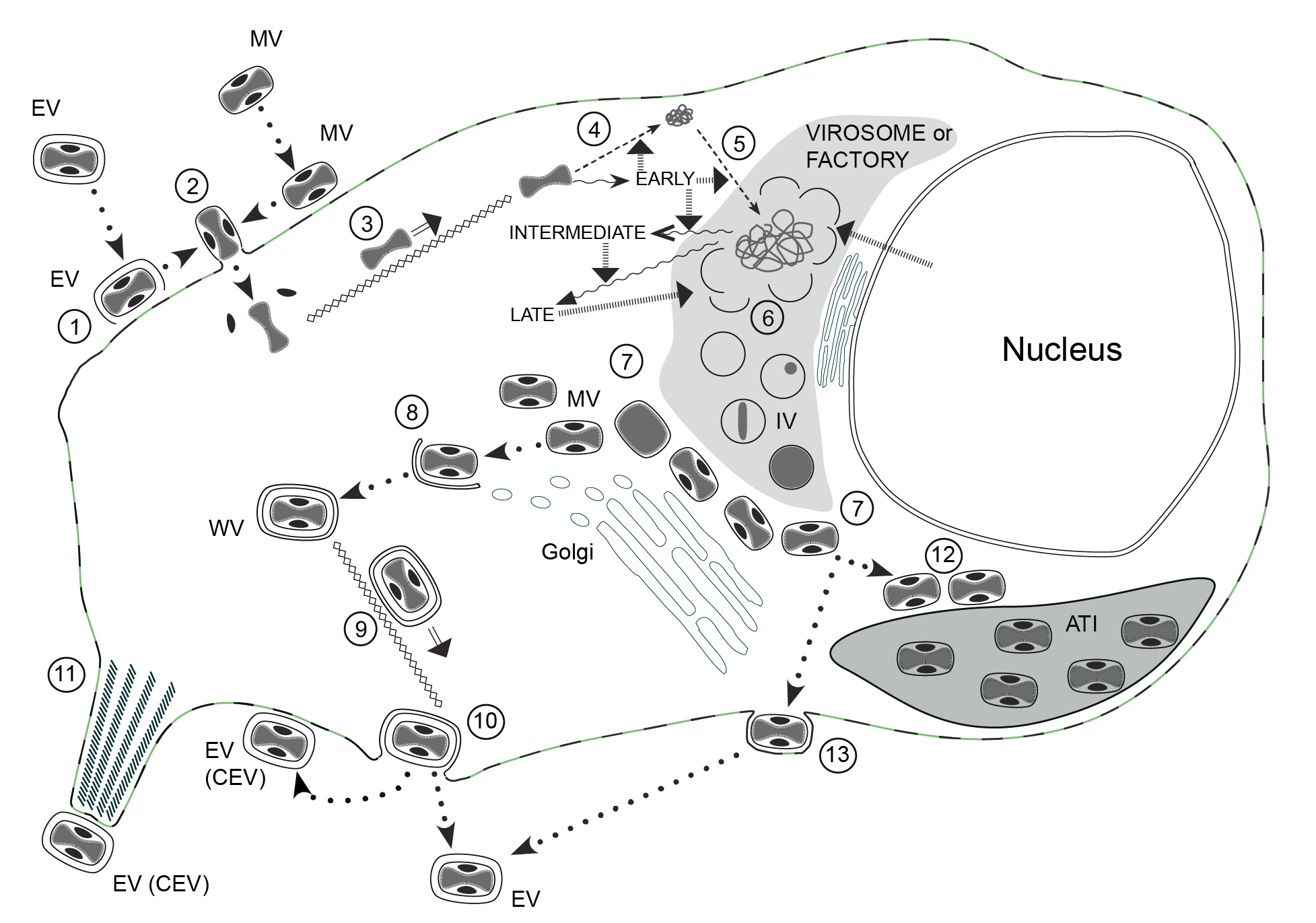 |
| Figure 3.Poxviridae The infectious cycle of poxviruses. The infectious cycle of poxviruses, based primarily on that of vaccinia virus (VACV): ATI, A-type inclusion body; IV, immature virion; MV, mature virion; WV, wrapped virion; EV, enveloped virion; CEV, cell-associated enveloped virion. See text for full details. (1) Disruption of envelope of EV upon binding to cell surface receptors, essentially revealing MV, which like naked MV can (2) fuse directly with the cell membrane (mediated by the fusion complex) to release the naked core (and lateral bodies). The core is (3) transported to the perinuclear region along microtubules. Early genes are expressed (wavy arrows) directly from the intact core; early gene products mediate: (4) uncoating of the core, (5) DNA replication and intermediate gene expression. Intermediate gene products (with involvement of some host proteins derived from the nucleus) mediate late gene expression. Late gene products include structural proteins, proteins required for morphogenesis and factors packed in the virion, such as the DNA polymerase required for early gene expression. Single membrane crescents are assembled (6) to enclose viral core proteins and genomic DNA (the latter is cleaved from concatameric intermediates), forming IV. These mature (7) to MV that, in VACV and many other mammalian poxviruses, are transported to the trans-Golgi/endosomal compartment for (8) wrapping with a double membrane to produce WV. These are (9) transported to the cell surface along microtubules, where they (10) exocytose, losing the outer of the two additional membranes, to form EV. The EV can remain on the cell surface as CEV or become free in the medium. CEV can (11) be propelled away from the cell on the tips of actin-driven projections. MV of some poxviruses can (12) alternatively be transported to and incorporated into ATI. The avipoxviruses do not appear to form WV to any significant extent, rather production of EV involves MV transport to the plasma membrane where they undergo budding to exit the cell (13). |
The entry of poxviruses into mammalian cells is divided into two phases: attachment of the virions to the cell surface and a fusion/entry event that delivers the viral core into the cellular cytoplasm (Moss 2006). The attachment phase is mediated largely by electrostatic interactions between the virion and cell surface moieties, particularly glycosaminoglycans and laminin, whereas fusion of the viral membrane with cellular membranes is mediated by a multi-subunit viral entry/fusion complex, comprising at least a dozen highly conserved viral proteins. Although EV differ from MV by possessing an extra membrane (the envelope), it is believed that the EV membrane is disrupted shortly after binding to the cell surface such that the entry/fusion stage is similar for MV and EV.
Polyadenylated, capped primary mRNA transcripts, representing about 50% of the genome, are initially synthesized from both DNA strands by enzymes within the core, including a virus-encoded multi-subunit RNA polymerase; transcripts are extruded from the core for translation. During synthesis of early proteins, host macromolecular synthesis is inhibited. Virus reproduction ensues in the host cell cytoplasm, producing basophilic (B-type) inclusions termed “viroplasms” or “virus factories”. The genome contains closely spaced protein-encoding ORFs, lacking introns, some of which may partially overlap. These ORFs are preceded by virus-specific promoters that temporally regulate transcription of three classes of mRNA. One class, the early genes, are expressed from partially uncoated virions prior to DNA replication (these encode many non-structural proteins, including enzymes involved in replicating the genome and modifying DNA and RNA, and proteins whose role is to neutralize the host response). Early genes also encode intermediate transcription factors. Intermediate genes, which encode late transcription factors, are expressed during the period of DNA replication and are required for subsequent late gene transcription. Finally, late genes are expressed during the post-replicative phase (these mainly encode virion structural proteins but also early transcription factors). Despite a cytoplasmic site of replication, there is evidence for the requirement of host nuclear proteins in post-replicative transcription. The mRNAs are capped, polyadenylated at the 3′ termini, and not spliced. Many intermediate, late and some early mRNAs have 5′-poly(A) tracts, that precede the encoded mRNA. Early protein synthesis is generally decreased during the transition to late gene expression, but some genes are expressed from promoters with both early and late activities. Certain proteins are modified post-translationally (e.g. by proteolytic cleavage, phosphorylation, glycosylation, ribosylation, sulphation, acylation, palmitylation and myristylation). Proteolytic cleavage of late proteins is required for virion morphogenesis.
The replication of the DNA genome appears to be mainly through the action of viral enzymes (Greseth and Traktman 2022). One model has DNA replication being initiated with the introduction of a single stranded nick near one (or both) of the terminal hairpins. The hairpin is unfolded and copied to the terminus. The two strands are separated at the terminus and hairpins reform, allowing nascent DNA to be extended along the whole length of the genome, through the opposite hairpin and back along the opposite strand, forming a concatemeric product. Resolution of the concatemers for packaging involves at least three virus-encoded functions: a nicking-joining DNase, topoisomerase I and a Holliday junction resolvase.
Genetic recombination between viruses in the same genus has been shown, and may occur between daughter molecules during replication (Evans 2022). Non-genetic genome reactivation generating infectious virus has been shown within, and between, members of genera in the subfamily Chordopoxvirinae, and forms the basis for the recovery of recombinant vaccinia virus from full-length genomic DNA (in, for instance, bacterial artificial chromosome vectors) by a helper virus (which can be removed by passage of the progeny through cells non-permissive for the helper, eg., fowlpox virus and passage in mammalian cells).
Virus morphogenesis begins following DNA replication and expression of early, intermediate and late genes. Particle assembly is initiated with the formation of crescent-shaped membrane structures in the intermediate compartment between the endoplasmic reticulum and the trans-Golgi network. Replicated, concatameric DNA is resolved into unit genomes and packaged, forming virion particles that mature into fully infectious MV (IMV). Some MV acquire an additional double layer of intracellular membrane (derived from the early endosomes or the trans-Golgi network) that contain unique virus proteins, to form WV (IEV). These WV are transported, by association with the cellular microtubule network, to the periphery of the cell where fusion with the plasma membrane ultimately results in release of EV (CEV and EEV). While both MV and EV are infectious, the external antigens on the two virion forms are different, and during infection the two virion types probably bind to different cellular receptors before uptake by mechanisms described above (Roberts and Smith 2008). Virus DNA and several proteins are organized as a nucleoprotein complex within the core of all infectious virions. The MV (IMV) contains an encompassing surface membrane, lateral bodies and the nucleoprotein core complex. For viruses within certain genera, negative-staining indicates that the core assumes a biconcave shape, apparently due to the large lateral bodies. Although the internal structure of some virions is revealed in thin sections, the detailed internal structure of parapoxvirus particles, for example is less evident. In negatively-stained preparations of parapoxviruses, sciuripoxviruses and the crocodylidpoxviruses superimposition of dorsal and ventral views of the surface filament normally produces a distinctive “criss-cross” surface appearance.
During natural infections, the virus is probably spread within an animal by EV (IEV and CEV) or through the movement of infected cells. An in vitro study has shown that infected cells, even before they have assembled infectious virions, can repel superinfecting virions and form actin-driven cellular projections (“actin tails” or “actin rockets”) that can further propel the superinfecting virion towards uninfected target cells, thereby rapidly spreading the infection.
Biology
Transmission of various members of the subfamily Chordopoxvirinae occurs by (1) droplet/aerosol, (2) direct contact, (3) arthropods (via mechanical means), or (4) indirect contact via fomites; transmission of members of the subfamily Entomopoxvirinae occurs between arthropods by mechanical means. Host range may be broad in laboratory animals and in tissue culture; however, in nature it is generally narrow. Many poxviruses of vertebrates produce dermal maculopapular, vesicular rashes after systemic or localized infections. Poxviruses infecting humans are zoonotic except for molluscum contagiosum virus (MCV) and the orthopoxvirus variola virus (VARV) (the etiologic agent of smallpox, now eradicated). Members may or may not be occluded within proteinaceous inclusions (subfamily Chordopoxvirinae: acidophilic (A-type) inclusion bodies, or subfamily Entomopoxvirinae: occlusions or spheroids). Occlusions may protect such poxviruses in environments where transmission possibilities are limited. Neutralizing antibodies and cell-mediated immunity play a major role in clearance of some vertebrate poxvirus infections. Reinfection rates are generally low and usually less severe. Molluscum contagiosum infections may recur, especially by autoinoculation of other areas of the skin with virus derived from the original lesions (e.g., by scratching).
Antigenicity
Within members of each genus of the subfamily Chordopoxvirinae there is considerable serologic cross-reactivity and cross-protection. Neutralizing antibodies are genus-specific. The nucleoprotein antigen, obtained by treatment of virus suspensions with 0.04 M NaOH and 56 °C treatment of virus suspensions, is highly cross-reactive among members. Orthopoxviruses have haemagglutinin antigens, although this is rare in other genera.
Subfamily demarcation criteria
Members of the subfamily Chordopoxvirinae infect vertebrates while members of the subfamily Entomopoxvirinae infect insects. Generally, gene content and synteny vary between the subfamilies, with only 49 gene products conserved between the two subfamilies.
Derivation of names
Avipoxvirus: from the Latin avis, meaning “bird”.
Capripoxvirus: from the Latin caper, meaning “goat”.
Centapoxvirus: from Central Africa, the geographical region in which yokapox virus, a member of the genus, was found.
Cervidpoxvirus: from the Latin cervus, meaning “deer”.
Chordopoxvirinae: from the phylum Chordata encompassing the vertebrates.
Crocodylidpoxvirus: from the Greek krokodilos, meaning “crocodile”.
Entomopoxvirinae: from the Greek entomon, meaning “insect”.
Leporipoxvirus: from the Latin lepus, meaning “hare”.
Macropopoxvirus: from the Latin macropod, meaning “large footed”.
Molluscipoxvirus: from the Latin molluscum, meaning “clam”, “snail”; referring to appearance of lesions produced by molluscum contagiosum virus, a member of the genus.
Mustelpoxvirus: from the Latin mustela, meaning “weasel”.
Orf virus: from the Scottish word orf based on the Icelandic word hrufa, both meaning “scab”, “boil”.
Orthopoxvirus: from the Greek orthos, meaning “straight”.
Oryzopoxvirus: from the genus (Oryzomys) of rodents in which members of the single species Cotia virus were found.
Parapoxvirus: from the Greek para, meaning “beside”.
Poxviridae: from the disease pox, itself from the Old English poc, pocc, meaning “pustule”.
Pteropopoxvirus: from the Greek pteropus, meaning “wing-footed”
Salmonpoxvirus: from salmon, itself from the Latin salmo, meaning “to leap”
Sciuripoxvirus: from the Latin sciurus, meaning “squirrel”
Suipoxvirus: from the Latin sus, meaning “swine”.
Vespertilionpoxvirus: from the Latin vespertilio, meaning “bat”
Yatapoxvirus: from Yaba monkey tumor virus and tanapox virus.
Most chordopoxvirus species names consist of two parts:
1. As prefix, a term describing the host from which the poxvirus is normally isolated and, as a suffix, the term “pox”. The prefix should be relevant in nature and scale to the taxonomic entity that best represents the relevant host taxon.
2. The word “virus”.
In some cases, intervening terminology describes a clinical feature of the disease caused by the virus. This nomenclature is normally used for poxviruses that cause characteristic pock-like skin lesions.
For those viruses that do not produce such lesions the following nomenclature would normally be used:
1. A term describing the host from which the poxvirus is normally isolated; it should be relevant in nature and scale to the taxonomic entity that best represents the relevant host taxon.
2. The word “poxvirus” (preferred), though “virus” is also used.
In some case, intervening terminology describes a clinical feature of the disease caused by the virus.
For the entomopoxviruses, the formal taxonomic name (genus, species) of the host precedes the term “entomopoxvirus”.
Please note that this naming convention will be updated to reflect the ICTV binomial naming requirements once the new binomial species names have been ratified.
Relationships within the family
Members of the family Poxviridae share a characteristic virion morphology, with subtle differences between some genera. Use of serology, nucleic acid hybridization and restriction enzyme fragment polymorphism are generally limited to within-genus studies. The two subfamilies represent the fundamental difference in hosts, based on whether or not they are chordate. The subfamilies were divided into genera based on host, disease (and in vitro biological characteristics), lack of extensive inter-genus antigenic cross-reactivity and phylogenetic analyses of limited numbers of genes. Genomic sequence analysis has supported traditional classification, though it is apparent that the divergence between the avian viruses is probably greater than would be accommodated within a single genus as indicated in the phylogenetic tree inferred at the family level (Figure 4.Poxviridae). A phylogenetic tree is also provided for exemplar viruses belonging to species in the genus Orthopoxvirus. (Figure 5.Poxviridae). Poxvirus phylogenetic reconstruction, especially at the family level, requires that amino acid multiple sequence alignments be used rather than nucleotide sequence alignments. This is due to the significant bias in nucleotide composition observed when comparing the genomic sequences of viruses from different genera. The genome GC content ranges from 18% in viruses belonging to the genus Betaentomopoxvirus to 65% or 66% for viruses in the genera Molluscipoxvirus and Sciuripoxvirus respectively (Lefkowitz et al., 2006).
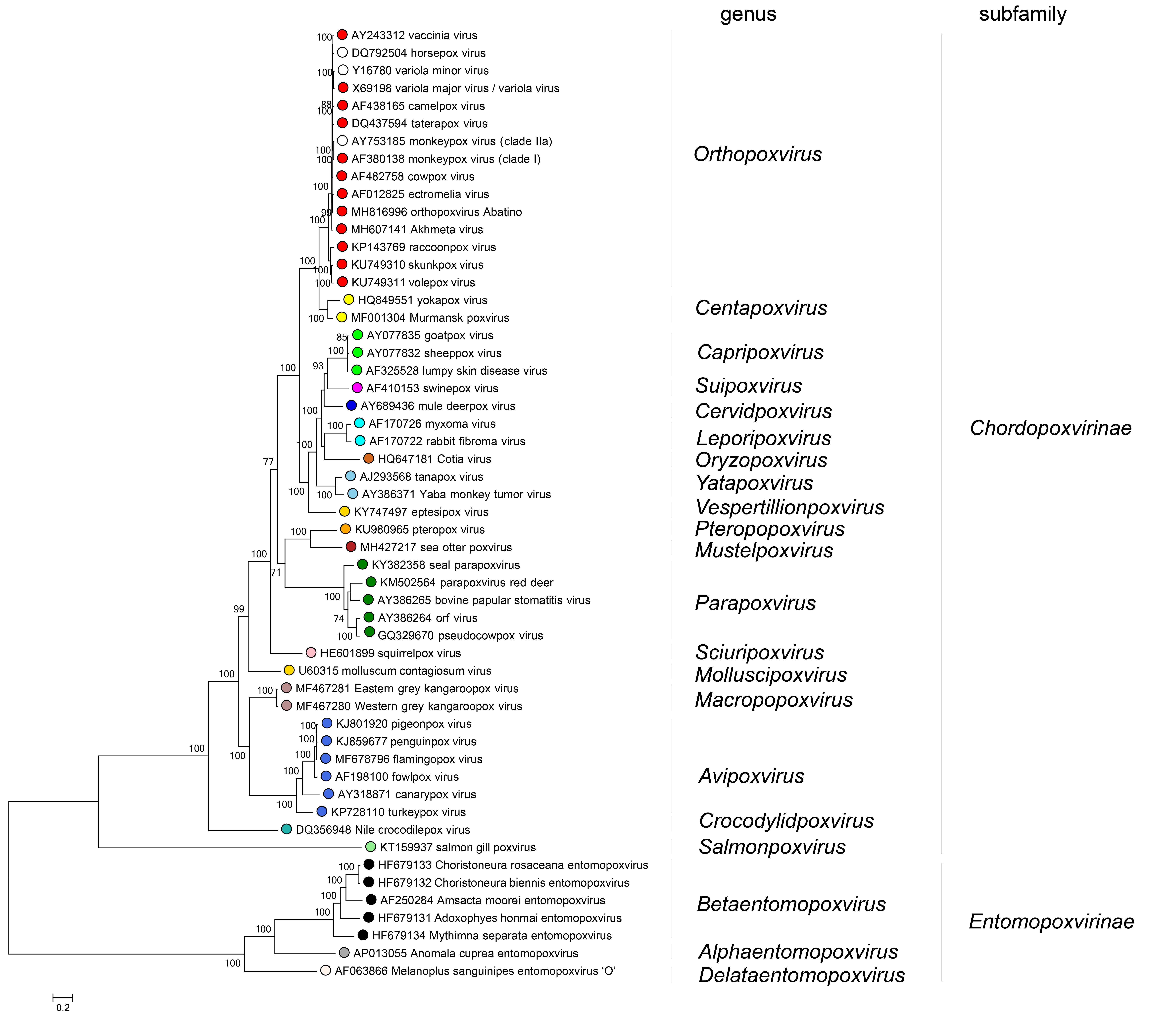 |
| Figure 4.Poxviridae. Phylogenetic relationships in the family Poxviridae. Representative viruses belonging to species in the family Poxviridae are shown based on their inferred phylogenetic placement. Individual genera in the subfamilies Chordopoxvirinae (vertebrate-infecting viruses) and Entomopoxvirinae (invertebrate-infecting viruses) are shown. Each species is represented by an exemplar isolate labeled with the name of the virus isolate and GenBank accession number for the complete genomic nucleotide sequence. Phylogeny was inferred using Maximum Likelihood (ML) phylogenetic inference using an amino acid multiple sequence alignment of 25 genes conserved between poxviruses. (Table 2.Poxviridae) The amino acid sequences for each gene were aligned using MUSCLE, and then the aligned sequences were concatenated into a single sequence for each isolate prior to phylogenetic inference. The program modeltest-ng (https://github.com/ddarriba/modeltest) was used to determine the optimum evolutionary model for phylogenetic reconstruction. Maximum Likelihood (ML) trees were inferred with the program raxml-ng (https://github.com/amkozlov/raxml-ng) using the model LG+I+G4+F, and 1,000 bootstrap replicates. |
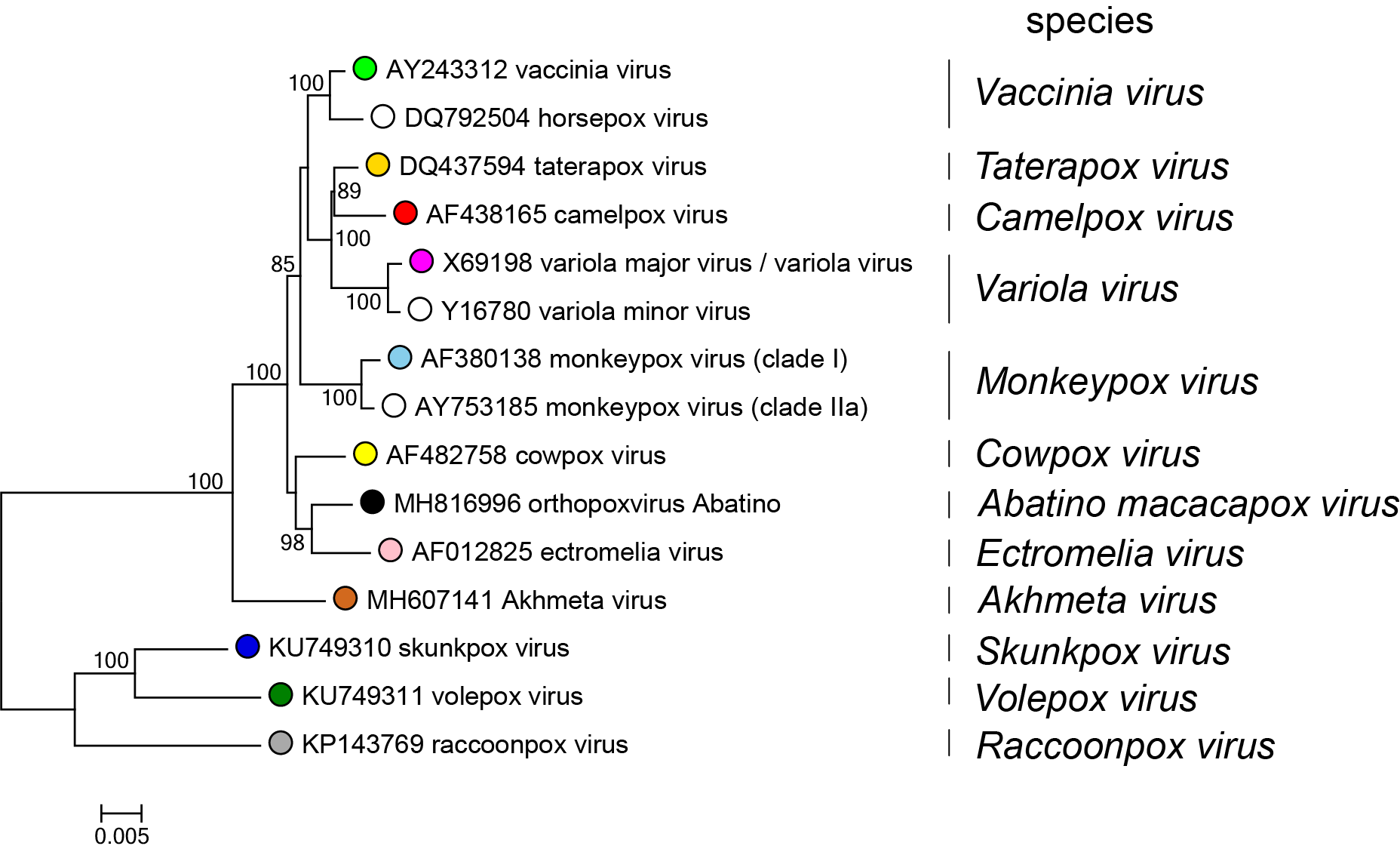 |
| Figure 5.Poxviridae Phylogenetic relationships in the genus Orthopoxvirius. Representative viruses belonging to species in the genus Orthopoxvirus are shown based on their inferred phylogenetic placement. Each species is represented by an exemplar isolate labeled with the name of the virus isolate and GenBank accession number for the complete genomic nucleotide sequence. Phylogeny was inferred using Maximum Likelihood (ML) phylogenetic inference using an amino acid multiple sequence alignment of 25 genes conserved between poxviruses. (Table 2.Poxviridae) The amino acid sequences for each gene were aligned using MUSCLE, and then the aligned sequences were concatenated into a single sequence for each isolate prior to phylogenetic inference. The program modeltest-ng (https://github.com/ddarriba/modeltest) was used to determine the optimum evolutionary model for phylogenetic reconstruction. Maximum Likelihood (ML) trees were inferred with the program raxml-ng (https://github.com/amkozlov/raxml-ng) using the model JTT+I+G4+F, and 1,000 bootstrap replicates. Bootstrap support is indicated where this was >70%. This tree shows a single, monophyletic branch for cowpox virus, but when all known genomic sequences characterized as “cowpox virus” are included in alignments and phylogenetic inference, multiple, polyphyletic cowpox virus branches are observed. (Antwerpen et al., 2019). Therefore, additional analysis needs to be performed on the sequences of these viruses to determine their phylogenetic placement and appropriate classification into species. |
Table 2.Poxviridae. Genes utilized for the analysis shown in Figure 4.Poxviridae and Figure 5.Poxviridae.
| Vaccinia virus Copenhagen (M35027) Gene designation | Protein name |
| A18 | DNA helicase |
| A2 | VLTF-3 |
| A22 | holliday junction resolvase |
| A23 | VITF-3 |
| A24 | RPO132 |
| A28 | IMV-MP |
| A3 | P4b |
| A32 | ATPase DNA packaging protein |
| A7 | VETF-L |
| D1 | large capping enzyme |
| D11 | NPH-I |
| D12 | small capping enzyme |
| D5 | NTPase |
| D6 | VETF-S |
| E1 | polyA polymerase large |
| E10 | sulfhydryl oxidase |
| E9 | DNA polymerase |
| G5 | FEN1 |
| H2 | IMV entry-fusion |
| H4 | RAP94 |
| H6 | DNA topoisomerase |
| I8 | NPH-II |
| J3 | polyA polymerase small |
| J6 | RPO147 |
| L1 | IMV membrane protein |
Relationships with other taxa
Members of the family Poxviridae are considered to be Nucleo-cytoplasmic large DNA viruses (NCLDV) that either replicate fully within the cytoplasm of host cells or start the process within the nucleus of the cell, but complete their replicative cycle within the cytoplasm. Phylogenetic analyses with 41 proteins conserved across the NCLDVs places the poxviruses in a clade with members of the family Asfaviridae, with a sister clade containing members of the families Iridoviridae, Phycodnaviridae and Mimiviridae (Iyer et al., 2006).

