Family: Rudiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Rudivirus
Type species Sulfolobus islandicus rod-shaped virus 2
Virion properties
Morphology
Virion has a stiff rod shape and measures about 600–900×23 nm (Figure 1). It is not enveloped and consists of a tube-like superhelix formed by dsDNA and multiple copies of a major structural protein. At each end, the tube carries plugs, about 50×6 nm, to which three tail fibers are anchored (Figure 1). These tail fibers appear to be involved in adsorption onto the host cell surface. The length of the virions is proportional to the size of the packaged viral DNA (Table 1).
Table 1 Properties of the rudiviruses
| Name | Origin | Virion length (nm) | Genome size (bp) | Total ORFs | G+C(%) | ITR length (bp) |
| SRV | Portugal | 702 | 28097 | 37 | 29.3 | 1030 |
| ARV1 | Italy | 610 | 24655 | 41 | 39.1 | 1365 |
| SIRV1 | Iceland | 830 | 32308 | 45 | 25.3 | 2032 |
| SIRV2 | Iceland | 900 | 35498 | 54 | 25.2 | 1626 |
Physicochemical and physical properties
The buoyant density of virions is 1.36 g cm−3. Virions are highly thermostable; autoclaving at 120 °C for at least 50 min is required for their inactivation. Virions are not inactivated by treatment with 6M urea, 0.1% Triton X-100, absolute ethanol and 2-octanol, but can be degraded by prolonged treatment with 0.1% SDS at 50 °C. Virions can act as a template for site-selective and spatially controlled chemical modification. Both the ends and the body of the virus, or the ends only, can be chemically addressed.
Nucleic acid
Genome is a single molecule of linear dsDNA, which ranges from 24,655 to 35,482 bp (Table 1).
Proteins
Virions contain one major highly glycosylated protein of about 14.5 kDa, and three minor proteins with molecular masses of about 50, 58 and 110 kDa, the largest of which is involved in formation of the terminal filaments. The major structural protein is shown to generate long tubular structures in vitro. The protein structure adopts a four-helix bundle fold that is stabilized by an extensive hydrophobic core, with helices ranging from 11 to 19 amino acids in length, and is identical to the four-helix bundle fold of the two major structural proteins of the Acidianus filamentous virus 1, a member of the family Lipothrixviridae.
Lipids
No lipid was detected.
Carbohydrates
None reported.
Genome organization and replication
The two strands of the linear DNA are covalently linked at both ends of the genome. The genomes carry long inverted terminal repeats, ranging from 1365 bp to 2032 bp (Table 1), which include multiple direct repeats. Although the sequences of the inverted terminal repeats are different for different rudiviruses, they all carry the 21 bp sequence AATTTAGGAATTTAGGAATTT near the genome ends, which may be an important signal for DNA replication. The finding of head-to-head and tail-to-tail linked replicative intermediates suggests a self-priming replication model.
The genome sequence and composition of the Acidianus and Stygiolobus rudiviruses differ significantly from those of the two Sulfolobus rudiviruses (Figure 2). A dUTPase and a Holliday junction resolvase are encoded in the virus genomes, and those of the Sulfolobus rod-shaped virus 2 have been functionally characterized as recombinant proteins in vitro. At least 10% of the encoded proteins are predicted to have different DNA-binding motifs and are presumed to be transcriptional regulators. One of these, protein SvtR of Sufolobus rod-shaped virus 2, with the structure similar to that of bacterial RHH proteins, was characterized in detail and shown to strongly repress the transcription of the minor structural protein. The transcriptional patterns of the rudiviruses are relatively simple, with few temporal expression differences.
Antigenic properties
No information is available.
Biological properties
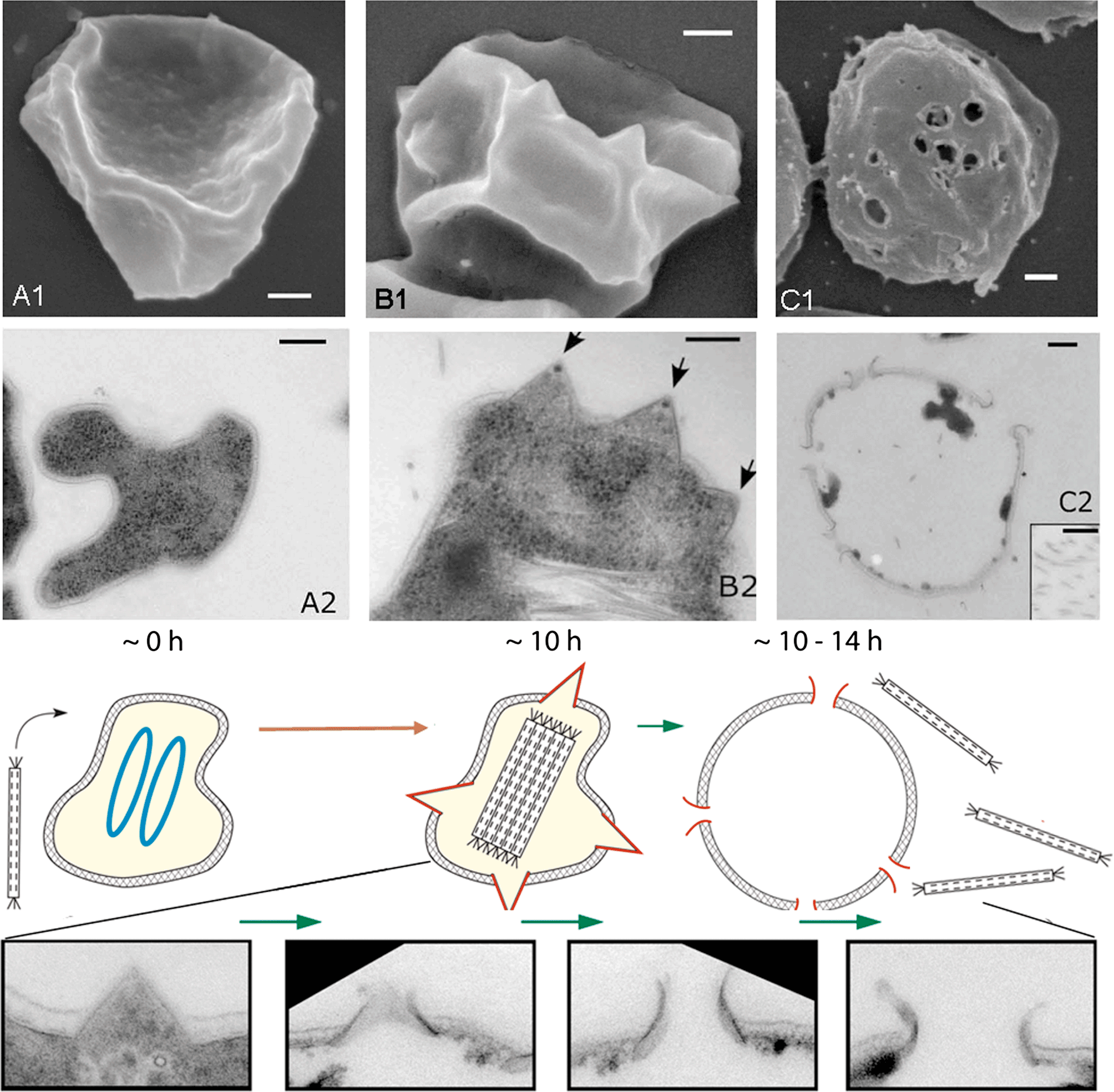
The viruses were isolated from extreme acidic geothermal environments, with temperatures above 80 °C and pH values below 3, of Iceland, Portugal and Italy. The hosts are members of the hyperthermophilic archaeal genera Sulfolobus, Acidianus and Stygiolobus. The viral genome does not integrate into the host chromosome. Sulfolobus islandicus rod-shaped virus 2 kills the host cell as a consequence of elaborated, well-orchestrated mechanism. Following virus infection, massive degradation of the host chromosomes occurs accompanied by formation of pyramidal structures on the host cell surface that rupture the S-layer (Figure 3). Close to the end of eclipse phase, the pyramidal structures open outwards, and create apertures through which mature virions escape the cell (Figure 3). SIRV2-encoded protein P98 is the major constituent of these exceptional cellular ultrastructures. This mechanism of virus release is unique and differs from lysis and egress systems of known bacterial and eukaryotic viruses.
Species demarcation criteria in the genus
Species in the genus differ in virion size, host range, size and nucleotide sequence of the genome.
List of species in the genus Rudivirus
| Sulfolobus islandicus rod-shaped virus 1 |
|
|
| Sulfolobus islandicus rod-shaped virus 1 | [AJ414696] | (SIRV1) |
| Sulfolobus islandicus rod-shaped virus 2 |
|
|
| Sulfolobus islandicus rod-shaped virus 2 | [AJ344259] | (SIRV2) |
| Acidianus rod-shaped virus 1 |
|
|
| Acidianus rod-shaped virus 1 | [AJ875026] | (ARV1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Rudivirus but have not been approved as species
| Stygiolobus rod-shaped virus | [FM164764] | (SRV) |
List of unassigned species in the family Rudiviridae
None reported.
Phylogenetic relationships within the family
Not available.
Similarity with other taxa
Originally, the two families of dsDNA viruses with linear genomes, the Rudiviridae and the Lipothrixviridae, were distinguished by differences in virion structure and this was later supported by comparative genomics. Nevertheless, a substantial fraction of orthologous genes, including some encoding glycosyl transferases and transcriptional regulators, are shared by the rudiviruses and lipothrixviruses. For example, of the 45 predicted genes of the rudivirus Sulfolobus islandicus rod-shaped virus 1, nine share orthologs with the lipothrixvirus Sulfolobus islandicus filamentous virus. Moreover, the structure of the major virion protein of the rudiviruses turned out to be identical to the structures of the two major virion proteins of the lipothrixvirus Acidianus filamentous virus 1. These observations indicate that the known linear dsDNA viruses, all of which infect hyperthermopilic members of the domain Archaea, share a common ancestor. One can suggest a sequence of evolutionary events in which the major virion protein of a “simpler” non-enveloped virion of the Rudiviridae has been duplicated and evolved so as to facilitate interactions with a hydrophobic envelope, producing the more complex virion of the Lipothrixviridae. Therefore it has been suggested that the families Rudiviridae and Lipothrixviridae could become members of a new viral order.
Derivation of name
Rudi: from Latin rudis, “small rod”.
Further reading
Blum, H., Zillig, W., Mallok, S., Domdey, H. and Prangishvili, D. (2001). The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology, 281, 6-9.
Bize, A., Karlsson, E.A., Ekefjärd, K., Quax, T. E. F., Pina, M., Prevost, M.-C., Forterre, P., Tenaillon, O., Bernander, R. and Prangishvili, D. (2009). A unique virus release mechanism in the Archaea. Proc. Natl Acad. Sci., USA, 106, 21155-21160.
Kessler, A., Brinkman, A.B., van der Oost, J. and Prangishvili, D. (2004). Transcription of the Rod-Shaped Viruses SIRV1 and SIRV2 of the Hyperthermophilic Archaeon Sulfolobus. J. Bacteriol., 186, 7745-7753.
Peng, X., Blum, H., She, Q., Mallok, S., Brugger, K., Garrett, R.A., Zillig, W. and Prangishvili, D. (2001). Sequences and replication of genomes of the archaeal rudiviruses SIRV-1 and SIRV-2: relationships to the archaeal Lipothrixvirus SIFV and some eukaryal viruses. Virology, 291, 226-234.
Prangishvili, D., Arnold, H.P., Gotz, D., Ziese, U., Holz, I., Kristjansson, J.K. and Zillig, W. (1999). A novel virus family, the Rudiviridae: Structure, virus–host interactions and genome variability of the sulfolobus viruses SIRV-1 and SIRV-2. Genetics, 152, 1387-1396.
Prangishvili, D., Forterre, P. and Garrett, R. A. (2006). Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol., 4, 837-848.
Prangishvili, D., Garrett, R.A. and Koonin, E.V. (2006). Evolutionary genomics of archaeal viruses: Unique viral genomes in the third domain of life. Virus Res., 117, 52-67.
Vestergaard, G., Häring, M., Peng, X., Rachel, R., Garrett, R.A. and Prangishvili, D. (2005). A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology, 336, 83-92.
Vestergaard, G., Shah, S.A., Bize, A., Reitberger, W., Reuter, M., Phan, H., Briegel, A., Rachel, R., Garrett, R.A. and Prangishvili, D. (2008). Stygiolobus rod-shaped virus and the interplay of crenarchaeal rudiviruses with the CRISPR antiviral system. J. Bacteriol., 190, 6837-6845.
Zillig, W., Prangishvili, D., Schleper, C., Elferink, M., Holz, I., Albers, S., Janekovic, D. and Götz, D. (1996). Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol. Rev., 18, 225-236.
Contributed by
Prangishvili, D.
Figures
Figure 1 (Left panel) Negative contrast electron micrographs of virions of Stygiolobus rod-shaped virus. The bars correspond to 200 nm. (Right panels) Electron tomography image of the virion: horizontal slice (0.7) showing the accumulated stain in the central part, and visualization of the data using Amira software. With an arrow is indicated a point of attachment of the three tail fibers. The scale bar represents 50 nm.
(Modified from Vestergaard et al. (2008). J. Bacteriol., 190, 6837-6845.)
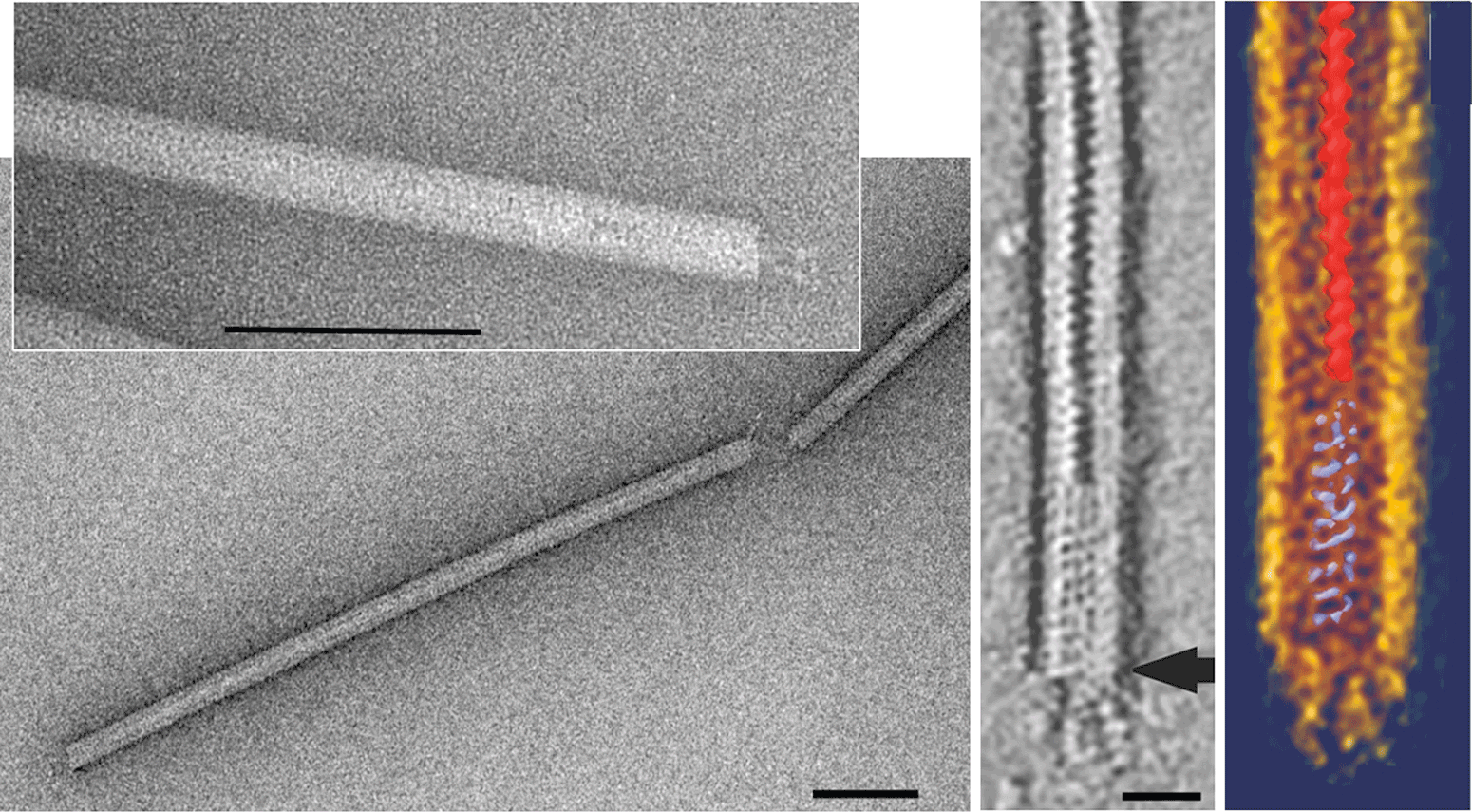
Figure 2 Genome organization of Stygiolobus rod-shaped virus (SRV), Sulfolopbus islandicus rod-shaped virus 1 (SIRV1) and Acidianus rod-shaped virus 1 (ARV1), showing the predicted ORFs and the ITRs (bold lines). SRV ORFs are identified by their amino acid lengths. Homologous genes shared between the rudiviruses are color-coded. Genes above the horizontal line are transcribed from left to right, and those below the line are transcribed in the opposite direction. Predicted functions or structural characteristics of the gene products are indicated as follows: sp, structural protein; rhh, ribbon-helix-helix protein; wh, winged helix protein; tm, transmembrane; tgt, tRNA guanine transglycosylase; hjh; Holliday junction helicase; hjr, Holliday junction resolvase; n, nuclease; du, dUTPase; ts, thymidylate synthase; sm, S-adenosylmethionine-dependent methyltransferase; gt, glycosyl transferase.
(From Vestergaard et al. (2008). J. Bacteriol. 190, 6837-6845.)
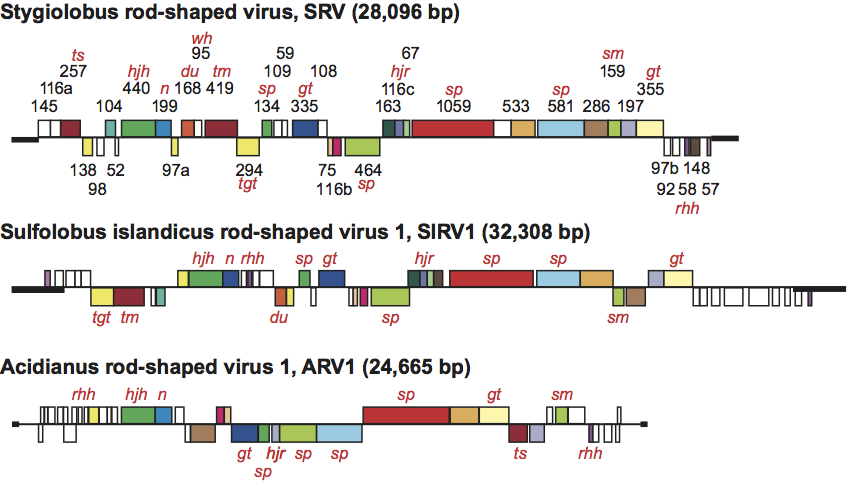
Figure 3 Transformation of Sulfolobus host cells following infection with Sulfolobus islandicus rod-shaped virus 2: (A1, A2), uninfected cells; (B1, B2), cells 10 h post-infection; (C1,C2), cells 1014 h post-infection. (A1, B1, C1) scanning electron micrographs; (A2, B2, C2) transmission electron micrographs of thin sections of negatively stained cells. Scale bars: 200 nm. The electron micrographs are accompanied by the schematic representation of the cell at different stages of the virus life cycle. Times after infection are indicated in hours. At 0 h, two chromosomes of the Sulfolobus host are shown in blue. Later they degrade concomitantly with formation of virus-induced pyramids (shown in red) and the intracellular clusters of assembled virions. Finally, at time points between 10 and 14 h, the virus-induced pyramids open (their remains are shown in red), the cell lyses and the virions are extruded. The gradual opening out of the virus-induced pyramids (at time points between 10 and 14 h) is illustrated in more details with fragments from electron micrographs of thin sections of the infected cell.
(Modified from Bize et al., 2009.)