Family: Secoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are non-enveloped 25–30 nm in diameter and exhibit icosahedral symmetry (T=1, pseudo T=3, Figure 1). Many virus preparations contain empty virus particles. In the case of viruses with a bipartite genome, the two RNAs are encapsidated in separate virions.
Physicochemical and physical properties
Different classes of virions are distinguished according to their buoyant densities (top, middle and bottom components, also termed T, M and B, Figure 1). The main virions (M and B components) contain RNA. Viruses belonging to the genera Sequivirus and Waikavirus, which have a single large monopartite genome, sediment with S20,W values of 150–190S. For viruses with a bipartite genome, virions containing RNA1 (B component) sediment at 110–135S. Virions containing RNA2 (M component) sediment at 84–128S and contain one or two molecules of RNA2. In cases where the size of RNA1 and RNA2 are similar, the M and B components may be difficult to separate. Empty shells (T component) sediment with S20,W values of 49–63S depending on the virus considered.
Nucleic acid
The genome consists of one or two molecules of linear positive sense ssRNA. The size of RNA(s) differs among genera (Table 1). The genomic RNA(s) contain a 3’-terminal poly(A) tract of variable length. The only known exception is the genomic RNA of a sequivirus (parsnip yellow fleck virus), which is apparently not polyadenylated. For several comoviruses and nepoviruses and for strawberry latent ringspot virus, an unassigned member of the family, a polypeptide, designated VPg (2–4 kDa) has been shown to be covalently bound at the 5′ end. The presence of a 5′-linked VPg has not been confirmed for other genera but has been suggested because, in many cases, infectivity of the RNA(s) has been shown to be protease-sensitive.
Table 1 Sizes of the genomes (nts) of representative viruses in the family Secoviridae
| Genus/virus* | RNA-1 | RNA-2 |
| Comovirus | 5,850–6,100 | 3,300–4,000 |
| Cowpea mosaic virus-SB | (5,889) | (3,810) |
| Fabavirus | 5,800–6,000 | 3,300–4,000 |
| Broad bean wilt virus 2-ME | (5,951) | (3,607) |
| Nepovirus | 7,200–8,400 | 3,700–7,300 |
| Grapevine fanleaf virus-F13 | (7,342) | (3,774) |
| Beet ringspot virus-S | (7,356) | (4,662) |
| Tomato ringspot virus-Rasp2 | (8,214) | (7,273) |
| Cheravirus | 6,800–7,100 | 3,200–3,700 |
| Cherry rasp leaf virus-USA | (6,992) | (3,274) |
| Sadwavirus | 6,800–7,000 | 5,300–5,600 |
| Satsuma dwarf virus-S58 | (6,795) | (5,345) |
| Torradovirus | 7,200–7,800 | 5,300–5,900 |
| Tomato torrado virus-PRI-0301 | (7,793) | (5,389) |
| Sequivirus | 9,800–10,000 |
|
| Parsnip yellow fleck virus-P121 | (9,871) | NA |
| Waikavirus | 11,800–12,500 | NA |
| Rice tungro spherical virus-LB | (12,433) |
|
| Unassigned species in the family |
|
|
| Strawberry latent ringspot virus-MEN454 | (7,496) | (3,842) |
| Strawberry mottle virus-1134 | (7,036) | (5,619) |
NA: not applicable.
* Please refer to tables within the text for the sequence accession numbers for the type isolate for each virus.
Proteins
Nepoviruses have a single coat protein (CP) of 52–60 kDa. Comoviruses, fabaviruses, sadwaviruses and strawberry latent ringspot virus have two CPs of 40–45 kDa and 21–29 kDa. Cheraviruses, torradoviruses, sequiviruses and waikaviruses have three CPs of similar sizes (24–35 kDa, 20–26 kDa and 20–25 kDa). The size and number of the CP(s) of two unassigned members of the family (strawberry mottle virus and black raspberry necrosis virus) has not been determined yet. Virions have 60 copies of each CP per particle. For three comoviruses (cowpea mosaic virus, bean pod mottle virus and red clover mottle virus) and one nepovirus (tobacco ringspot virus), the atomic structure has been solved and found to be very similar (pseudo T=3) to that of viruses belonging to the family Picornaviridae. Each capsid subunit is made of three beta-barrels (jelly roll domains) that can be present in one large CP with three jelly roll domains (Nepovirus), two CPs (one large CP including two jelly roll domains and one smaller CP with a single jelly roll domain; Comovirus, Fabavirus and Sadwavirus) or three CPs each containing a single jelly roll domain (Cheravirus, Torradovirus, Sequivirus, Waikavirus) (Figure 2).
Lipids
None reported.
Carbohydrates
None reported. A report that the CPs of comoviruses contain carbohydrates has later been shown to be mistaken.
Genome organization and replication
Unfractionated viral RNA is highly infective. In the case of viruses with a bipartite genome, neither RNA species alone can infect plants systemically. RNA1 carries all the information required for replication and can replicate in individual cells in the absence of RNA2 although no virus particles are produced (as demonstrated for comoviruses and nepoviruses).
Viral proteins are usually expressed as large polyproteins, which are cleaved by 3C-like proteinases. Each RNA usually encodes a single polyprotein (Figure 3). A notable exception is the RNA2 of torradoviruses, which contains two open reading frames. Another exception is the RNA2 of comoviruses. Although a single large ORF is present, internal initiation at a second AUG allows the formation of two distinct polyproteins. The 3′ UTR of the RNA varies in length, sometimes even within a genus (nepoviruses). In some cases, extensive regions of sequence identity between RNA1 and RNA2 are found in the 5′ and/or 3′ UTRs (Figure 3).
Within the polyproteins, protein domains are organized in a manner that is common to that of other members of the order Picornavirales (Figure 3). The replication block contains domains characteristic of NTP-binding proteins (NTB or putative helicase), 3C-like proteinase (Pro) and RNA-dependent RNA polymerase (Pol). In viruses with a monopartite genome, the structural proteins are located upstream of the replication block in the single polyprotein. In viruses with a bipartite genome, structural proteins are contained in the RNA2-encoded polyprotein. In comoviruses, cheraviruses and nepoviruses, the movement protein is located upstream of the CP(s), and enables viral movement to adjacent cells. Both movement protein and CP(s) are required for cell-to-cell movement of the virus. The movement protein of comoviruses and nepoviruses was shown to be a structural component of tubular structures that traverse the cell wall and contain virus-like particles. Putative movement proteins have been suggested to be encoded upstream of the CP(s) coding regions for many other viruses in the family but their biological function has not been confirmed.
The RNA1-encoded 3C proteinase cleaves both RNA1 and RNA2-encoded polyproteins. The cleavage site specificity of the proteinase differs with the specific genera considered (and in the case of nepoviruses it differs with the specific subgroup, Table 2). An amino acid in the substrate-binding pocket of the proteinase interacts directly with the amino acid in the −1 position of the cleavage site and plays a key role in the specificity of the proteinase.
Formation of replication complexes has been studied for comoviruses and nepoviruses. Replication occurs in association with intracellular membranes derived from the endoplasmic reticulum. Two RNA1-encoded proteins (the NTB protein and the protein immediately upstream of NTB) interact directly with ER membranes and have been implicated in the proliferation of membrane vesicles in the cytoplasm of infected cells and in the assembly of the replication complex. This has not been studied for other viruses in the family.
Table 2 Cleavage site specificity of the 3C-like proteinase of viruses in the family Secoviridae
| Genus | Proteinase substrate binding pocket* | Dipeptide at cleavage site† |
| Comovirus | His | Q/G, Q/M, Q/S, Q/T, Q/A |
| Fabavirus | His | Q/S, Q/A, Q/G |
| Nepovirus |
|
|
| Subgroup A | Leu | R/G, C/S, C/A, A/S, G/E, G/V, C/G |
| Subgroup B | Leu | K/S, K/A, R/A, R/S, R/G |
| Subgroup C | His | Q/G, Q/S, D/S |
| Cheravirus | His | Q/G, E/G |
| Sadwavirus | ? | R/G, T/S, T/N, A/N, A/S, A/A |
| Torradovirus | His | ? |
| Sequivirus | Leu | ? |
| Waikavirus | His | Q/S, Q/M, Q/V, Q/A |
| Unassigned species in the family |
|
|
| Strawberry latent ringspot virus | His | S/G |
| Strawberry mottle virus | His | Q/G |
* The indicated amino acid in the substrate-binding pocket of the proteinase interacts with the amino acid at the −1 position of the cleavage site and plays an important role in determining the cleavage site specificity of the proteinase. In the case of sadwaviruses, neither a His nor a Leu are present at the equivalent position in the deduced amino acid sequence of the proteinase.
† Cleaved dipeptides at the cleavage sites are shown with the scissile bond indicated with the slanted line. The amino acids are shown using the one-letter code. Proteolytic cleavages at dipeptides shown in red have been confirmed experimentally. Dipeptides shown in black are putative cleavage sites, inferred from sequence alignments.
Antigenic properties
Virus preparations are usually good immunogens and polyclonal antibodies prepared against purified virus particles recognize all CPs. Species belonging to the same genus can be serologically interrelated, but often distantly.
Biological properties
All members of the family infect plants. Host range and symptoms vary with the genera and viruses considered (Table 3). Many viruses in the family have a known biological vector, although some (sequiviruses) require a helper virus and others do not have a known vector. Most viruses are transmissible experimentally by mechanical inoculation. However, waikaviruses are not sap-transmissible. Many viruses are readily transmissible by seed or pollen.
Table 3 Biological properties of viruses in the family Secoviridae
| Genus | Host range | Vector | Seed or pollen transmission |
| Comovirus | Narrow (Leguminosae) | Beetle | Rare |
| Fabavirus | Wide | Aphid | Rare |
| Nepovirus | Wide | Nematode (most) or mite (blackcurrant reversion virus) or unknown | Seed and/or pollen |
| Cheravirus | Wide or narrow | Nematode (cherry rasp leaf virus) or unknown | Seed |
| Sadwavirus | Wide | Unknown | Seed |
| Torradovirus | Narrow | Whitefly | Unlikely |
| Sequivirus | Relatively wide | Aphid (requires helper virus) | None |
| Waikavirus | Narrow | Aphid or leafhopper | None |
| Unassigned species |
|
|
|
| Strawberry latent ringspot virus | Wide | Nematode | Seed |
| Strawberry mottle virus | Relatively wide | Aphid | None |
Genus demarcation criteria in the family
- Number of genomic RNAs
- Number of protein domains and/or processing sites within the polyprotein(s)
- Number of CPs
- Presence of additional ORFs and/or subgenomic RNAs
- Clustering as a single branch in phylogenetic trees derived from amino acid sequence alignments of the conserved Pro-Pol region when compared with other genera of the family Secoviridae (see Figure 5 below). The Pro-Pol region is delineated by the “CG” motif of the 3C-like proteinase and the “GDD” motif of the polymerase. Identification of proteinase cleavage sites is not required to delineate the Pro-Pol region.
Not all criteria may need to be met simultaneously.
Species demarcation criteria
- CP aa sequence with less than 75% identity (for viruses with two or three CPs, combined CP sequences are considered)
- Conserved Pro-Pol region aa sequence (as defined above) with less than 80% identity
- Differences in antigenic reactions
- Distinct host range
- Distinct vector specificity
- Absence of cross-protection
- For viruses with a bipartite genome, absence of re-assortment between RNA1 and RNA2.
Not all criteria need to be met simultaneously. In some cases, sequence information alone can be a good indicator of a distinct species (i.e., when the percentage of sequence identity in both the Pro-Pol and CP(s) regions is well below the proposed cut-off). However, analyzing only one region of the genome is generally not sufficient and both the Pro-Pol and CP(s) regions should be considered. In cases where the percentage of sequence identity in one or both sequences is near the proposed cut-off (e.g., between 75 and 85% in the Pro-Pol region or between 70 and 80% in the CP(s) region), other criteria should be considered and information on biological properties of the virus (host range, vector specificity, possibility of reassortment between RNAs) is useful. For example, beet ringspot virus (BRSV) and tomato black ring virus (TBRV) (genus Nepovirus) are closely related in the Pro-Pol sequence (89% sequence identity) but are much more divergent in the CP sequence (62% sequence identity). They differ in their antigenic reactions and also in the specificity of nematode transmission (BRSV is transmitted more efficiently by Longidorus elongatus and TBRV is transmitted more efficiently by Longidorus attenuatus).
Subfamily Comovirinae
Distinguishing features
The genome of members of the subfamily Comovirinae consists of two ssRNAs with a 5′-bound polypeptide (VPg) and a 3′ poly(A) tail. Members of the subfamily group as a single branch in phylogenetic trees using the conserved Pro-Pol region (see section on phylogenetic relationships in the family and Figure 5 below). Other genera are more distantly related. Within the subfamily, genera are distinguished by their specific genomic organization, biological properties and phylogenetic relations. Each genus within the subfamily Comovirinae represents a single sub-branch in the Pro-Pol phylogenetic tree.
Genus Comovirus
Type species Cowpea mosaic virus
Distinguishing features
The comovirus capsid is made of two types of polypeptides (large CP: 40–45 kDa and small CP: 21–27 kDa). The small CP suppresses RNA silencing and surface-exposed amino acids are required for this function.
The 5′ and 3′ NTRs of RNA-1 and RNA-2 are similar in sequence but not identical. RNA-2 is translated into two largely overlapping polyproteins that are processed into three domains. Production of the smaller polyprotein is caused by internal initiation at a downstream AUG, which is placed in a more favorable context than the upstream AUG (Figure 4). The 58K protein released from the N-terminus of the larger polyprotein (P2) is necessary for replication of RNA-2. The 48K protein released from the N-terminus of the smaller polyprotein (P2′) is the MP, with a typical “LPL” motif. The CP domains are encoded at the C-terminus of both polyproteins. The MP and the CPs are required for cell-to-cell movement of the virus. The MP is a structural component of tubular structures containing virus-like particles that traverse the cell wall. The C-terminal region of the MP also interacts with the large CP. RNA-1 is translated into a single polyprotein that is processed into five domains, through alternative processing pathways (Figure 4). The N-terminal 32K protein limits the processing of the RNA-1-encoded polyprotein in cis and assists the processing of the RNA-2-encoded polyprotein. This protein is often referred to as the protease co-factor or Co-Pro. The replication block on the RNA1-encoded polyprotein includes the 58K protein with sequence motifs characteristic of an NTP-binding helicase, the VPg, the Pro and the Pol. The 32K Co-Pro and 58K NTB proteins are involved in inducing the cytopathic structure through proliferation of ER-derived membranes.
Comoviruses have narrow host ranges, 11 of the 15 species being restricted to a few species of the family Leguminosae. Mosaic and mottle symptoms are characteristic, but usually not ringspots. Transmission in nature is exclusively by beetles, especially members of the family Chrysomelidae. Beetles retain their ability to transmit virus for days or weeks.
List of species in the genus Comovirus
|
| [RNA1] | [RNA2] |
|
| Andean potato mottle virus |
|
|
|
| Andean potato mottle virus-C |
| [L16239] | (APMoV-C) |
| Bean pod mottle virus |
|
|
|
| Bean pod mottle virus - KentuckyG7 | [U70866 = NC_003496] | [M62738 = NC_003495] | (BPMV-KenG7) |
| Bean pod mottle virus - K-Hopkins1 | [AF394608] | [AF394609] | (BPMV-KHop) |
| Bean pod mottle virus-K-Hopkins2 | [DQ139274*] |
|
|
| Bean pod mottle virus - K-Hancock1 | [AF394606] | [AF394607] | (BPMV-KHan) |
| Bean pod mottle virus-IL-Carbondale 1 | [AY744931] | [AY744933] | (BPMV-IL-Cb1) |
| Bean pod mottle virus-IL-Carbondale 1-sugroup II | [AY744932] |
| (BPMV-IL-Cb1-II) |
| Bean rugose mosaic virus |
|
|
|
| Bean rugose mosaic virus-Parana |
| [AF263548*] | (BRMV-Parana) |
| Broad bean stain virus |
|
|
|
| Broad bean stain virus-Loewe-07013PC |
| [FJ028650] | (BBSV-07013PC) |
| Broad bean true mosaic virus |
|
|
|
| (Echtes Ackerbohnemosaik virus) |
|
|
|
| (Vicia virus 1) |
|
|
|
| Broad bean true mosaic virus-PV-0098 |
| [FJ442942*] | (BBTMV-PV-0098) |
| Cowpea mosaic virus |
|
|
|
| (Cowpea yellow mosaic virus) |
|
|
|
| Cowpea mosaic virus –SB | [X00206 = NC_003549] | [X00729 = NC_003550] | (CPMV-SB) |
| Cowpea severe mosaic virus |
|
|
|
| (Arkansas cowpea mosaic virus) |
|
|
|
| (Cowpea mosaic virus - severe) |
|
|
|
| (Trinidad cowpea mosaic virus) |
|
|
|
| (Puerto Rico cowpea mosaic virus) |
|
|
|
| Cowpea severe mosaic virus - DG | [M83830 = NC_003545] | [M83309 = NC_003544] | (CPSMV-DG) |
| Glycine mosaic virus |
|
|
|
| Glycine mosaic virus-New South Wales |
|
| (GMV-NSW) |
| Pea green mottle virus |
|
|
|
| Pea green mottle virus-Czechoslovakia |
|
| (PGMV-CZ) |
| Pea mild mosaic virus |
|
|
|
| Pea mild mosaic virus-New Zealand |
|
| (PMiMV-NZ) |
| Quail pea mosaic virus |
|
|
|
| (Bean curly dwarf mosaic virus) |
|
|
|
| Quail pea mosaic virus-Arkansas |
|
| (QPMV-AR) |
| Radish mosaic virus |
|
|
|
| (Radish enation mosaic virus) |
|
|
|
| Radish mosaic virus-Japan | [AB295643 = NC_010709] | [AB295644 = NC_010710] | (RaMV-Japan) |
| Radish mosaic virus-California | [AB456531] | [AB456532] | (RaMV-Cal) |
| Radish mosaic virus-CB3 |
| [FJ442945] | (RaMV-CB3) |
| Radish mosaic virus-CB9 |
| [FJ442946] | (RaMV-CP9) |
| Radish mosaic virus-RaMV1 | [EU450837] | [EU450838] | (RaMV-1) |
| Red clover mottle virus |
|
|
|
| Red clover mottle virus - S | [X64886 = NC_003741] | [M14913 = NC_003738] | (RCMV-S) |
| Red clover mottle virus-SI-9 |
| [FJ442940*] | (RCMV-SI-9) |
| Red clover mosaic virus-SI-12 |
| [FJ442941*] | (RCMV-SI-12) |
| Squash mosaic virus |
|
|
|
| (Cucurbit ring mosaic virus) |
|
|
|
| (Muskmelon mosaic virus) |
|
|
|
| (Pumpkin mosaic virus) |
|
|
|
| Squash mosaic virus - Y | [AB054688 = NC_003799] | [AB054689 = NC_003800] | (SqMV-Y) |
| Squash mosaic virus - Arizona |
| [AF059533] | (SqMV-Ari) |
| Squash mosaic virus - CH99/211 | [EU421059] | [EU421060] | (SqMV-CH99/211) |
| Squash mosaic virus - Kimble |
| [AF059532] | (SqMV-Kim) |
| Squash mosaic virus - Melon |
| [M96148*] | (SqMV-Melon) |
| Ullucus virus C |
|
|
|
| Ullucus virus C-Andes |
|
| (UVC-Andes) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.Only one type isolate is listed for each species. The type isolate was chosen as the isolate with the most complete nucleotide sequence information. When several isolates with complete sequence information are available, the isolate with the earlier database deposition is provided as the type isolate.
* Sequences do not comprise the complete genome segment.
List of other related viruses which may be members of the genus Comovirus but have not been approved as species
|
| [RNA1] | [RNA2] |
|
| Turnip ringspot virus-Toledo | [FJ712026=NC_013218] | [FJ712027=NC_013219] | (TuRSV-Toledo) |
Turnip ringspot virus (TuRSV) is related to radish mosaic virus (RaMV). They infect similar hosts. The degree of aa sequence identity between the two viruses is close to the proposed species demarcation criteria (73% aa sequence identity in the combined CP region and 80% aa sequence identity in the Pro-Pol region). It is not known whether re-assortment between the RNAs of TuRSV and RaMV is possible. Therefore, the taxonomic position of TuRSV as a distinct species in the genus Comovirus or as a distant strain of the species Radish mosaic virus remains unclear.
Genus Fabavirus
Type species Broad bean wilt virus 1
Distinguishing features
The genomic organization of fabaviruses is similar to that of comoviruses, although it is not known whether RNA2 encodes two overlapping polyproteins in fabaviruses (Figure 3). The cleavage of polyproteins is presumed to be similar to that of comoviruses but this has not been investigated in detail. Fabaviruses have wide host ranges among dicotyledonous plants and some families of monocotyledonous plants. Symptoms are ringspots, mottling, mosaic, distortion, wilting and apical necrosis. In nature, fabaviruses are transmitted by aphids in a non-persistent manner.
List of species in the genus Fabavirus
|
| [RNA1] | [RNA2] |
|
| Broad bean wilt virus 1 |
|
|
|
| (Nasturtium ringspot virus) |
|
|
|
| Broad bean wilt virus 1-ATCC PV132 | [AB084450 = NC_005289] | [AB084451 = NC_005290] | (BBWV1-PV132) |
| Broad bean wilt virus 1-ATCC PV176 |
| [AB018703*] | (BBWV1-PV176) |
| Broad bean wilt virus 1-Ben | [AY781171] | [AY781172] | (BBWV1-Ben) |
| Broad bean wilt virus 1-Japan |
| [AB110537] | (BBWV1-Japan) |
| Broad bean wilt virus 1-Singapore |
| [AF225955] | (BBWV1-Singapore) |
| Broad bean wilt virus 2 |
|
|
|
| (Parsley virus 3) |
|
|
|
| (Petunia ringspot virus) |
|
|
|
| (Plantago II virus) |
|
|
|
| Broad bean wilt virus 2 - ME | [AF225953 = NC_003003] | [AF225954 = NC_003004] | (BBWV2-ME) |
| Broad bean wilt virus 2- 1-2 |
| [AB-18701*] | (BBWV2-1-2) |
| Broad bean wilt virus 2-3-10 |
| [AB161177*] | (BBWV2-3-10) |
| Broad bean wilt virus 2 - B935 | [AF149425] |
| (BBWV2-B935) |
| Broad bean wilt virus 2-BC | [FJ485684*] | [FJ485685*] | (BBWV2-BC) |
| Broad bean wilt virus 2 - China |
| [AJ132844] | (BBWV2-China) |
| Broad bean wilt virus 2-E |
| [AB018699*] | (BBWV2-E) |
| Broad bean wilt virus 2-fruit sage |
| [EF392660*] | (BBWV2-fruit sage) |
| Broad bean wilt virus 2 - IA | [AB051386] | [AB032403] | (BBWV2-IA) |
| Broad bean wilt virus 2 - IP | [AB023484] | [AB018698] | (BBWV2-IP) |
| Broad bean wilt virus 2-JK |
| [AB161178*] | (BBWV2-JK) |
| Broad bean wilt virus 2-JP92V2 |
| [AB076671*] | (BBWV2-JP92V2) |
| Broad bean wilt virus 2 - Korea K |
| [AF104335] | (BBWV2-Kor) |
| Broad bean wilt virus 2-L |
| [AB018700*] | (BBWV2-L) |
| Broad bean wilt virus 2 - MB7 | [AB013615] | [AB013616] | (BBWV2-MB7) |
| Broad bean wilt virus 2-Nagaimo |
| [AB207244*] | (BBWV2-Nagaimo) |
| Broad bean wilt virus 2 - P158 |
| [AF228423] | (BBWV2-P158) |
| Broad bean wilt virus 2 - PV131 | [BBU65984*] | [U65985] | (BBWV2-PV131) |
| Broad bean wilt virus 2-RS |
| [AB261176*] | (BBWV2-RS) |
| Broad bean wilt virus 2-Rehmannia |
| [GQ202215] | (BBWV2-Rehmannia) |
| Broad bean wilt virus 2-South Korea | [AF144234] |
| (BBWV2-SK) |
| Broad bean wilt virus 2-SP |
| [AB161179*] | (BBWV2-SP) |
| Patchouli mild mosaic virus | [AB050782 = NC_003975] | [AB011007 = NC_003974] | (PatMMV) |
| Gentian mosaic virus |
|
|
|
| Mikania micrantha mosaic virus | [EU158250 = NC_011190] | [EU158249 = NC_011189] | (MMMV) |
| Gentian mosaic virus-AFV1 |
| [AB076672*] | (GeMV-AFV1) |
| Gentian mosaic virus-N-1 | [AB084452] | [AB084453] | (GeMV-N-1) |
| Lamium mild mosaic virus |
|
|
|
| Lamium mild mosaic virus-Cambridge |
|
| (LMMV-Cambridge) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Fabavirus but have not been approved as species
None reported.
Genus Nepovirus
Type species Tobacco ringspot virus
Distinguishing features
Nepoviruses are the only known members of the family that encode a single large CP of 52–60 kDa. Genome organization and expression are similar to those of comoviruses, except that RNA-2 specifies a single polyprotein of 105–207 kDa. Nepoviruses can be divided into three subgroups. Subgroup A has an RNA-2 of 3,700–4,000 nts in length, present in both M and B components. Subgroup B has an RNA-2 of 4,400–4,700 nts in length, present only in the M component. Subgroup C has an RNA-2 of 6,400–7,300 nts in length, present in M component particles that are sometimes barely separable from those of B component. The three subgroups also differ in the cleavage sites recognized by their proteinase (see Table 2).
Additional linear or circular satellite RNAs, which sometimes modulate symptoms, are found associated with several nepoviruses of all three subgroups. They are either linear (1100–1800 nt) with a 5′-linked VPg, a 3′ poly(A) tail and encoding a 36–48 kDa polypeptide, or circular (300–460 nt) and apparently non-coding. They are present in some natural isolates but are not necessary for virus accumulation.
The RNA2-encoded polyprotein of subgroup A and B nepoviruses is processed into three domains. In grapevine fanleaf virus (GFLV), the N-terminal protein of the RNA2-encoded polyprotein (P2A) was shown to be involved in RNA-2 replication. The two other protein domains are the MP and the unique CP. Both are required for cell-to-cell movement of the virus. Similarly to comoviruses, the MP has a LPL motif, interacts with the CP and is a structural component of tubular structures containing virus-like particles and traversing the cell wall. In tomato ringspot virus (ToRSV) (subgroup C), the N-terminal region of the RNA2-encoded polyprotein is cleaved at an additional site, defining two domains (X3 and X4). The X3 protein contains some sequence similarity with the P2A protein of GFLV but the X4 protein is a unique protein of unknown function. The RNA-1 of nepoviruses is translated into a single polyprotein that is processed into six domains. The C-terminal region of the polyprotein contains the replication block, and is similar to that of comoviruses (NTB-VPg-Pro-Pol). In contrast, the N-terminal region of the polyprotein contains an additional cleavage site defining two protein domains (X1 and X2) instead of the single domain present upstream of NTB in the comovirus genome. Cleavage at this additional site was demonstrated for arabis mosaic virus (subgroup A) and ToRSV (subgroup C). A putative cleavage site at this position has been implied for other nepoviruses. The function of X1 is unknown. X2 contains a sequence motif in common with the comovirus Co-Pro protein but does not seem to modulate the activity of the proteinase. However, similarly to the comovirus Co-Pro, the X2 protein of ToRSV associates with ER-derived membranes and a role in viral replication has been proposed. When comparing RNA-1 and RNA-2, the 5′ and 3′ NTRs are similar in sequence but not identical in subgroup A nepoviruses. In subgroup B nepoviruses, the 5′-NTRs also show sequence similarity between RNA-1 and RNA-2, while the 3′-NTRs are identical in both RNAs. In subgroup C nepoviruses, both NTRs are identical or nearly identical between RNA-1 and RNA-2. The region of sequence similarity extends into part of the coding region of the polyproteins in ToRSV, but not in blackcurrant reversion virus.
Nepoviruses are widely distributed in temperate regions. The natural host range of nepoviruses varies from wide to restricted, depending on the virus. Ringspot symptoms are characteristic, but mottling and spotting are equally frequent. Twelve species are acquired and transmitted persistently by longidorid nematodes (Xiphinema, Longidorus or Paralongidorus spp.), three are transmitted by pollen, one is transmitted by mites (blackcurrant reversion virus) and the others have no known biological vector. Seed and/or pollen transmission is very common. In herbaceous plants, the symptoms induced by nepoviruses are often transient, with newly emerging leaves appearing symptomless a few weeks after infection (the so-called “recovery” phenomenon). Symptom recovery is associated with induction of RNA silencing, an antiviral defence, and is sometimes (but not always) accompanied with reduced concentration of viral RNAs.
List of species in the genus Nepovirus
|
| [RNA1] | [RNA2] |
| |
| SUBGROUP A |
|
|
| |
| Arabis mosaic virus |
|
|
| |
| (Ash ring and line pattern virus) |
|
|
| |
| (Forsythia yellow net virus) |
|
|
| |
| (Raspberry yellow dwarf virus) |
|
|
| |
| (Rhubarb mosaic virus) |
|
|
| |
| Arabis mosaic virus - NW | [AY303786 = NC_006057] | [AY017339 = NC_006056] | (ArMV-NW) | |
| Arabis mosaic virus-butterbur |
| [AB279740] | (ArMV-butterbur) | |
| Arabis mosaic virus-lilac |
| [D10086*] | (ArMV-lilac) | |
| Arabis mosaic virus-lily |
| [AB279741] | (ArMV-lily) | |
| Arabis mosaic virus-Lv | [EU617326] | [EU617327] | (ArMV-Lv) | |
| Arabis mosaic virus-MD |
| [EU433920*] | (ArMV-MD) | |
| Arabis mosaic virus-narcissus |
| [AB279740] | (ArMV-narcissus) | |
| Arabis mosaic virus - P2 |
| [X81814, X81815] | (ArMV-P2) | |
| Arabis mosaic virus-Ta |
| [EF426853] | (ArMV-Ta) | |
| Arracacha virus A |
|
|
| |
| Arracacha virus A-Huanuco |
|
| (AVA-Huanuco) | |
| Artichoke Aegean ringspot virus |
|
|
| |
| Artichoke Aegean ringspot virus |
|
| (AARSV) | |
| Cassava American latent virus |
|
|
| |
| Cassava American latent virus-South America |
|
| (CsALV-SA) | |
| Grapevine deformation virus |
|
|
| |
| Grapevine deformation virus-Turkey |
| [AY291208] | (GDeV-Turkey) | |
| Grapevine deformation virus-Turkey2 |
| [AY233975*] | (GDeV-Turkey2) | |
| Grapevine fanleaf virus |
|
|
| |
| (Grapevine infectious degeneration virus) |
|
|
| |
| Grapevine fanleaf virus - F13 | [D00915 = NC_003615] | [X16907 = NC_003623] | (GFLV-F13) | |
| Grapevine fanleaf virus - NW |
| [AY017338] | (GLFV-NW) | |
| Potato black ringspot virus |
|
|
| |
| Potato black ringspot virus - Greece | [AJ616715*] |
| (PBRSV-Gr) | |
| Tobacco ringspot virus - potato calico |
| [EU281548*] | (TRSV-calico) | |
| Raspberry ringspot virus |
|
|
| |
| (Raspberry Scottish leaf curl virus) |
|
|
| |
| (Redcurrant ringspot virus) |
|
|
| |
| Raspberry ringspot virus - Cherry | [AY303787 = NC_005266] | [AY303788 = NC_005267] | (RpRSV-Che) | |
| Raspberry ringspot virus - Grapevine | [AY310444] | [AY310445] | (RpRSV-Gra) | |
| Raspberry ringspot virus- Himalaya (E) |
| [AF110476, AF224695*] | (RpRSV-E) | |
| Raspberry ringspot virus-Lloyd George |
| [AF110795, AF224696*] | (RpRSV-Lloyd George) | |
| Raspberry ringspot virus-MX |
| [AF111114, AF224697*] | (RpRSV-MS) | |
| Raspberry ringspot virus-ORR |
| [AF116189, AF226157*] | (RpRSV-ORR) | |
| Raspberry ringspot virus- RAC815 | [DQ004849*] | [EF534293] | (RpRSV-RAC815) | |
| Raspberry ringspot virus-Shepherd |
| [AF116190, AF226158*] | (RpRSV-Shepherd) | |
| Raspberry ringspot virus-Tarvit |
| [AF116191 , AF226159*] | (RpRSV-Tarvit) | |
| Tobacco ringspot virus |
|
|
| |
| Tobacco ringspot virus-Bud blight | [U50869 = NC_005097] | [AY363727 = NC_005096] | (TRSV-Bud blight)) | |
| Tobacco ringspot virus-cherry-UK |
| [AF461163*] | (TRSV-cherry) | |
| Tobacco ringspot virus- Iran |
| [AF461164*] | (TRSV-Iran) | |
| Tobacco ringspot virus-SD1 |
| [AY787756*] | (TRSV-SD1) | |
| SUBGROUP B |
|
|
| |
| Artichoke Italian latent virus |
|
|
| |
| Artichoke Italian latent virus-Southern Italy |
| [X87254*] | (AILV-SI) | |
| Beet ringspot virus |
|
|
| |
| (Tomato black ring virus - Scottish) |
|
|
| |
| Beet ringspot virus-S | [D00322 = NC_003693] | [X04062 = NC_003694] | (BRSV-S) | |
| Cocoa necrosis virus |
|
|
| |
| (Cacao swollen shoot virus - S) |
|
|
| |
| Cocoa necrosis virus-ATCC PV-283 |
| [EU741694*] | (CoNV-PV283) | |
| Crimson clover latent virus |
|
|
| |
| Crimson clover latent virus-UK Herforshire |
|
| (CCLV-UK) | |
| Cycas necrotic stunt virus |
|
|
| |
| Cycas necrotic stunt virus-Japan | [AB073147 = NC_003791] | [AB073148 = NC_003792] | (CNSV-Japan) | |
| Cycas necrotic stunt virus-gladiolus |
| [AB237656*] | (CNSV-gladiolus) | |
| Grapevine Anatolian ringspot virus |
|
|
| |
| Grapevine Anatolian ringspot virus-Turkey |
| [AY291207] | (GARSV-Turkey) | |
| Grapevine chrome mosaic virus |
|
|
| |
| Grapevine chrome mosaic virus-Hungary | [X15346 = NC_003622] | [X15163 = NC_003621] | (GCMV-Hungary) | |
| Mulberry ringspot virus |
|
|
| |
| Mulberry ringspot virus- Japan |
|
| (MRSV-Japan) | |
| Olive latent ringspot virus |
|
|
| |
| Olive latent ringspot virus-Italy |
| [AJ277435] | (OLRSV-Italy) | |
| Tomato black ring virus |
|
|
| |
| (Bean ringspot virus) |
|
|
| |
| (Lettuce ringspot virus) |
|
|
| |
| (Potato bouquet virus) |
|
|
| |
| Tomato black ring virus - MJ | [AY157993 = NC_004439] | [AY157994 = NC_004440] | (TBRV-MJ) | |
| Tomato black ring virus - ED |
| [X80831] | (TBRV-ED) | |
| SUBGROUP C |
|
|
| |
| Apricot latent ringspot virus |
|
|
| |
| Apricot latent ringspot virus-Modesto |
| [AJ278875*] | (ALRSV-Modesto) | |
| Artichoke yellow ringspot virus |
|
|
| |
| (Tomato white ringspot virus) |
|
|
| |
| Artichoke yellow ringspot virus-Allium cepa | [AM087671*] |
| (AYRSV-Ac) | |
| Artichoke yellow ringspot virus-Cynara scolymus | [AM087673*] |
| (AYRSV-Cs) | |
| Artichoke yellow ringspot virus-Vicia faba | [AM087672*] |
| (AYRSV-Vf) | |
| Tomato white ringspot virus-T818 | [EF205130*] | [EF205131*] | (TWRSV-T818) | |
| Blackcurrant reversion virus |
|
|
| |
| Blackcurrant reversion virus-Finland Piikkio | [AF368272 = NC_003509] | [AF020051 = NC_003502] | (BRV-Piikkio) | |
| Blueberry leaf mottle virus |
|
|
| |
| Blueberry leaf mottle virus-Michigan | [U20622*] | [U20621*] | (BLMoV-Michigan) | |
| Cassava green mottle virus |
|
|
| |
| Cassava green mottle virus-Solomon Island |
|
| (CsGMV-Solomon) | |
| Cherry leaf roll virus |
|
|
| |
| (Elm mosaic virus) |
|
|
| |
| (Golden elderberry virus) |
|
|
| |
| (Walnut black line virus) |
|
|
| |
| Cherry leaf roll virus-walnut W8 | [Z34265*] | [U24694*] | (CLRV-W8) | |
| Chicory yellow mottle virus |
|
|
| |
| (Parsley carrot leaf virus) |
|
|
| |
| Chicory yellow mottle virus-Italy |
|
| (ChYMV-Italy) | |
| Grapevine Bulgarian latent virus |
|
|
| |
| Grapevine Bulgarian latent virus-Bulgaria |
|
| (GBLV-Bulgaria) | |
| Grapevine Tunisian ringspot virus |
|
|
| |
| Grapevine Tunisian ringspot virus-Tunisia |
|
| (GTRSV-Tunisia) | |
| Hibiscus latent ringspot virus |
|
|
| |
| Hibiscus latent ringspot virus-Ibadan |
|
| (HLRSV-Ibadan) | |
| Lucerne Australian latent virus |
|
|
| |
| Lucerne Australian latent virus-TN |
|
| (LALV-TN) | |
| Myrobalan latent ringspot virus |
|
|
| |
| Myrobalan latent ringspot virus-Prunus cerasifera |
|
| (MLRSV-Pc) | |
| Peach rosette mosaic virus |
|
|
| |
| (Grape decline virus) |
|
|
| |
| (Grapevine degeneration virus) |
|
|
| |
| Peach rosette mosaic virus-Michigan | [AF016626] |
| (PRMV-Michigan) | |
| Potato virus U |
|
|
| |
| Potato virus U-Lichte Industrie |
|
| (PVU-LI) | |
| Tomato ringspot virus |
|
|
| |
| (Grape yellow vein virus) |
|
|
| |
| (Nicotiana virus 13) |
|
|
| |
| (Peach yellow bud mosaic virus) |
|
|
| |
| (Tobacco ringspot virus n°2) |
|
|
| |
| Tomato ringspot virus-raspberry2 | [L19655 = NC_003840] | [D12477 = NC_003839] | (ToRSV-Rasp2) | |
| Tomato ringspot virus - Grape yellow vein | [AF135407*] | [AF135411*] | (ToRSV-GYV) | |
| Tomato ringspot virus- Peach yellow bud | [AF135408*] | [GQ325249, AF135412*] | (ToRSV-PYB) | |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome segment.
List of other related viruses which may be members of the genus Nepovirus but have not been approved as species
None reported.
Other genera in the family Secoviridae
Genus Cheravirus
Type species Cherry rasp leaf virus
Distinguishing features
Cheraviruses have three CPs of similar sizes. In some cases, these proteins are not fully or reproducibly resolved from each other by electrophoresis. The genome of cheraviruses is bipartite and the genomic organization is similar to that of comoviruses, although RNA2 is thought to encode a single polyprotein (Figure 3). The RNA2-encoded movement protein of apple latent spherical virus (ALSV) is 42 kDa, suggesting that translation initiation occurs at the second AUG, which is in a better context. Tubular structures containing virus-like particles are observed in infected cells and are likely involved in cell-to-cell movement of the virus. The movement protein and all three CPs are necessary for cell-to-cell movement of the virus. The MP binds to VP25, one of the three CPs. VP20 of ALSV, another CP, is a suppressor of silencing that interferes with systemic movement of the silencing signal.
The host range is broad or narrow, depending on viruses, and includes weed plants found in the vicinity of infected crops. Symptoms are usually mild or absent. Cherry rasp leaf virus is transmitted by nematodes (Xiphinema americanum) in the field, and is readily seed-transmitted. ALSV is also seed-transmitted through both embryo and pollen in apple.
List of species in the genus Cheravirus
|
| [RNA1] | [RNA2] |
|
| Apple latent spherical virus |
|
|
|
| Apple latent spherical virus-Fukushima | [AB030940 = NC_003787] | [AB030940 = NC_003788] | (ALSV-Fukushima) |
| Cherry rasp leaf virus |
|
|
|
| Cherry rasp leaf virus-USA potato | [AJ621357 = NC_006271] | [AJ621358 = NC_006272] | (CRLV-USA) |
| Flat apple virus | [AY764390] | [AY122330] | (FAV) |
| Stocky prune virus |
|
|
|
| Stocky prune virus-Brugères | [DQ143874*] | [DQ143875*] | (StPV- Brugères) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome segment.
List of other related viruses which may be members of the genus Cheravirus but have not been approved as species
None reported.
Genus Sadwavirus
Type species Satsuma dwarf virus
Distinguishing features
Similarly to comoviruses, sadwaviruses have two CPs, one large and one small. The genome of sadwaviruses is bipartite and the genomic organization is similar to that of comoviruses. The proteinase of sadwaviruses is distinct from that of other viruses in the family in that it does not have a conserved His or Leu in the active site. In addition, the cleavage sites recognized by sadwavirus proteinases are unique with an A or a T at the −1 position (Table 2). In contrast to comoviruses, there is no evidence that two overlapping polyproteins are encoded by RNA-2. Similarly to some nepoviruses, extensive sequence identity between RNA-1 and RNA-2 are found in the 5′ NTRs as well as in the 5′end of the putative coding region.
All isolates of satsuma dwarf virus infect citrus trees. There are no known biological vectors.
List of species in the genus Sadwavirus
|
| [RNA1] | [RNA2] |
|
| Satsuma dwarf virus |
|
|
|
| Satsuma dwarf virus-S58 | [AB009958 = NC_003785] | [AB009959 = NC_003786] | (SDV-S58) |
| Citrus mosaic virus-Az-1 |
| [AB032752*] | (CiMV-Az1) |
| Citrus mosaic virus-Ci-968 |
| [AB465581] | (CiMV-CI968) |
| Citrus mosaic virus-LB-1 |
| [AB032751*] | (CiMV-LB1) |
| Citrus mosaic virus-ND-1 |
| [AB465582] | (CiMV-ND1) |
| Natsudaidai dwarf virus |
| [AB032750] | (NDV) |
| Navel orange infectious mottling virus | [AB022887*] | [AB465583] | (NIMV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Sadwavirus but have not been approved as species
None reported.
Genus Torradovirus
Type species Tomato torrado virus
Distinguishing features
Similarly to cheraviruses, torradoviruses have a bipartite genome and three capsid proteins. The RNAs are polyadenylated. Presence of a VPg at the 5′ end of the RNAs has not been tested experimentally. The genomic organization is similar to that of other members of the family with a bipartite genome. The replication block is found in the RNA-1 polyprotein while the structural proteins are present in the C-terminal region of the RNA-2 polyprotein. A putative movement protein suggested upstream of the CP domains shares little sequence similarity with that of other bipartite members of the family with the exception of the small LPL motif. A distinguishing feature of the torradovirus genome is the presence of a second open reading frame upstream and partially overlapping with the large ORF in RNA-2 (Figure 3). This reading frame encodes a protein of unknown function, which exhibits a large degree of sequence diversity (61–74%) among torradoviruses. The 3′ NTRs share a large region with near sequence identity (>99%) between the RNA-1 and RNA-2 of a given species but differ substantially between species.
Tomato torrado virus was reported to be transmitted by whiteflies. Information is not available regarding vector transmission of other torradoviruses.
List of species in the genus Torradovirus
|
| [RNA1] | [RNA2] |
|
| Tomato torrado virus |
|
|
|
| Tomato torrado virus-PRI-0301 | [ DQ388879 = NC_009013] | [DQ388880 = NC_009032] | (ToTV-PRI-0301) |
| Tomato torrado virus-CE | [EU476181*] | [EU476182*] | (ToTV-CE) |
| Tomato torrado virus-Kra |
| [EU652402*] | (ToTV-Kra) |
| Tomato torrado virus-Ros |
| [EU652401*] | (ToTV-Ros) |
| Tomato torrado virus-Wal’03 | [EU563948] | [EU563947] | (ToTV-Wal’03) |
| Tomato torrado virus-W1 |
| EU090252* | (ToTV-W1) |
| Tomato torrado virus-W2 |
| EU090253* | (ToTV-W2) |
| Tomato torrado virus-CAN |
| EF436286* | (ToTYV-CAN |
| Tomato torrado virus-PAN1 |
| EU0934037* | (ToTV-PAN1 |
| Tomato torrado virus-H1 |
| FJ616996* | (ToTV-H1 |
| Tomato torrado virus-H2 |
| FJ616997* | (ToTV-H2 |
| Tomato torrado virus-H3 |
| FJ616998* | (ToTV-H3 |
| Tomato torrado virus-Sicily09 |
| GU903899* | (ToTV-Sicily09) |
| Tomato marchitez virus |
|
|
|
| (Tomato apex necrosis virus) |
|
|
|
| Tomato marchitez virus- PRI-0601 | [EF681764 = NC_010987] | [EF681765 = NC_010987] | (ToMarV-PRI-0601) |
| Tomato apex necrosis virus-VE434 | [EF063641*] | [EF063642*] | (ToANV-VE434) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Torradovirus but have not been approved as species
|
| [RNA1] | [RNA2] |
|
| Tomato chocolate virus-G01 | [FJ7560489] | [FJ560490] | (ToChV-G01) |
| Tomato chocolate spot virus-Guatemala | [GQ305131] | [GQ305132] | (ToChSV-Guatemala) |
Tomato chocolate virus (ToChV) and tomato chocolate spot virus (ToChSV) are related to tomato marchitez virus (ToMarV) and to a lesser degree to tomato torrado virus (ToTV). All viruses infect tomato and cause similar symptoms. Comparison of the aa sequence of the Pro-Pol and combined CP regions would suggest that ToChV and ToChSV are distant strains of ToMarV (82–90% aa sequence identity for the Pro-Pol region and 83–87% aa sequence identity in the combined CP region among the three viruses). However, other regions of the genome (RNA2-encoded ORF1 and the 3′ NTR) show significant sequence variation. In addition the length of the 3′ NTR varies significantly among these viruses. It is not known whether reassortment between the RNAs of ToMarV, ToChV and/or ToChSV is possible. Therefore, the taxonomic position of ToChV and ToChSV as two distinct species in the genus Torradovirus, as two strains of a single new species in the genus Torradovirus or as distant strains of the species Tomato marchitez virus remains unclear.
Genus Sequivirus
Type species Parsnip yellow fleck virus
Distinguishing features
Virions contain three CPs of about 32–34, 22–26 and 22–24 kDa. The genome consists of a single molecule of ssRNA that encodes a single large polyprotein. The replication block (NTB-Pro-Pol) is contained in the C-terminal region of the polyprotein. The structural protein domains are present in the N-terminal region of the polyprotein but are separated from the N-terminus by a protein domain of about 40–60 kDa. Infectivity of the genome is susceptible to proteinase treatment suggesting the presence of a 5′-linked VPg. The parsnip yellow fleck virus (PYFV) RNA is not polyadenylated. This is a unique property within this family. In contrast, the RNA of carrot necrotic dieback virus is polyadenylated. Tubular structures containing virus-like particles have been observed traversing the cell wall of PYFV-infected cells. However, their role in cell-to-cell movement has not been investigated and the presence of a movement protein in the polyprotein (possibly upstream of the CPs) needs to be confirmed.
The natural host range of sequiviruses includes species in several plant families. Transmission of PYFV is by aphids in a semi-persistent manner. However, it is dependent on the presence of a helper virus in the genus Waikavirus.
List of species in the genus Sequivirus
| Carrot necrotic dieback virus |
|
|
| Carrot necrotic dieback virus-Anthriscus | [EU980442] | (CNDV-Anthriscus) |
| Dandelion yellow mosaic virus |
|
|
| Dandelion yellow mosaic virus-DSM2 | [DQ675189*] | (DaYMV-DSM2) |
| Parsnip yellow fleck virus |
|
|
| (Celery yellow net virus) |
|
|
| Parsnip yellow fleck virus-P121 | [D14066=NC_003628] | (PYFV-P121) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Sequivirus but have not been approved as species
None reported.
Genus Waikavirus
Type species Rice tungro spherical virus
Distinguishing features
The genomic organization of waikaviruses is similar to that of sequiviruses. However, small ORFs have been identified near the 3′ end of the RNA or overlapping with the main polyprotein but in a different reading frame (Figure 3). Some experimental evidence has been presented suggesting that subgenomic RNAs are produced from the 3′ region of the RNA. The biological significance of the small open reading frames or of the putative subgenomic RNAs is not known. The genomic RNAs are polyadenylated at their 3′ end. The presence of a 5′-linked VPg has not been confirmed experimentally.
The natural host range of waikaviruses is usually restricted to species within a few plant families. Waikaviruses are not sap-transmitted. Field transmission is in the semi-persistent manner by aphids or leafhoppers. A virus-encoded helper protein is probably needed. Some waikaviruses are helper viruses for the insect transmission of other viruses: PYFV (genus Sequivirus) in the case of anthriscus yellows virus and rice tungro bacilliform virus (family Caulimoviridae) in the case of rice tungro spherical virus (this association being responsible for the very damaging rice tungro disease).
List of species in the genus Waikavirus
| Anthriscus yellows virus |
|
|
| Anthriscus yellows virus-anthriscus sylvestris |
| (AYV-As) |
| Maize chlorotic dwarf virus |
|
|
| Maize chlorotic dwarf virus-Tennessee | [U67839 = NC_003626] | (MCDV-Tennessee) |
| Maize chlorotic dwarf virus- Severe | [AY362551] |
|
| Maize chlorotic dwarf virus-M1 | [AY829112] | (MCDV-M1) |
| Rice tungro spherical virus |
|
|
| Rice tungro spherical virus-Los Banos Phillipine | [M95497 = NC_001632] | (RTSV-LB) |
| Rice tungro spherical virus-Andra Pradesh | [GQ227731*] | (RTSV-AP) |
| Rice tungro spherical virus-India | [X95284*} | (RTSV-India) |
| Rice tungro spherical virus-Malaysia | [U70989*} | (RTSV-Malaysia) |
| Rice tungro spherical virus-Orissa | [AM234048] | (RTSV-Orissa) |
| Rice tungro spherical virus-Phil1 | [U71440*] | (RTSV-Phil1) |
| Rice tungro spherical virus-Phil.2 | [X98396*] | (RTSV-Phil2) |
| Rice tungro spherical virus-RTV3 | [AF484118*] | (RTSV-RTV3) |
| Rice tungro spherical virus - Vt6 | [AB064963] | (RTSV-Vt6) |
| Rice tungro spherical virus-West Bengal | [AM234049} | (RTSV-West Bengal) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Waikavirus but have not been approved as species
None reported.
List of unassigned species in the family Secoviridae
|
| [RNA1] | [RNA2] |
|
| Black raspberry necrosis virus |
|
|
|
| Black raspberry necrosis virus-BrdAV-1 | [DQ344639 = NC_008182] | [DQ344640 = NC_008183] | (BRNV-BrdAV-1) |
| Black raspberry necrosis virus-GSMP | [EU419645} |
| (BRNV-GSMP) |
| Strawberry latent ringspot virus |
|
|
|
| (Rhubarb virus 5) |
|
|
|
| Strawberry latent ringspot virus-NCGR MEN 454.001 | [AY860978 = NC_006964] | [AY860979 = NC_006965] | (SLRSV-MEN454) |
| Strawberry latent ringspot virus-flowering cherry |
| [X75165*] | (SLRSV-cherry) |
| Strawberry latent ringspot virus-Hampshire |
| [X77466*] | (SLRSV-H) |
| Strawberry mottle virus |
|
|
|
| Strawberry mottle virus-1134 | [AJ311875 = NC_003445] | [AJ311876 = NC_003446] | (SMoV-1134) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Strawberry mottle virus and black raspberry virus are related to satsuma dwarf virus (SDV) in phylogenetic trees using the conserved Pro-Pol region (see Figure 5). They also have a bipartite genome. However, the nature of their capsid protein(s) and their genomic organization are not known. For this reason, they are considered unassigned species in the family Secoviridae. Strawberry latent ringspot virus was formerly considered a sadwavirus because it has two CPs and some distant relation with SDV in phylogenetic trees using the Pro-Pol sequence (Figure 5). However, its genomic organization is more related to that of cheraviruses (with the exception of the number of CPs, Figure 3) and it branches more closely with cheraviruses than with sadwaviruses in the phylogenetic trees using the Pro-Pol sequence (Figure 5). For these reasons, it is not considered a sadwavirus any more, and is now an unassigned species in the family Secoviridae.
Phylogenetic relationships within the family
Members of the family Secoviridae were previously classified in two different families: Comoviridae (including the genera Comovirus, Fabavirus and Nepovirus) and Sequiviridae (including the genera Sequivirus and Waikavirus) and in two unassigned genera: Cheravirus and Sadwavirus. The families and genera were recently amalgamated to create the new family Secoviridae, which regroups all plant picornavirales.
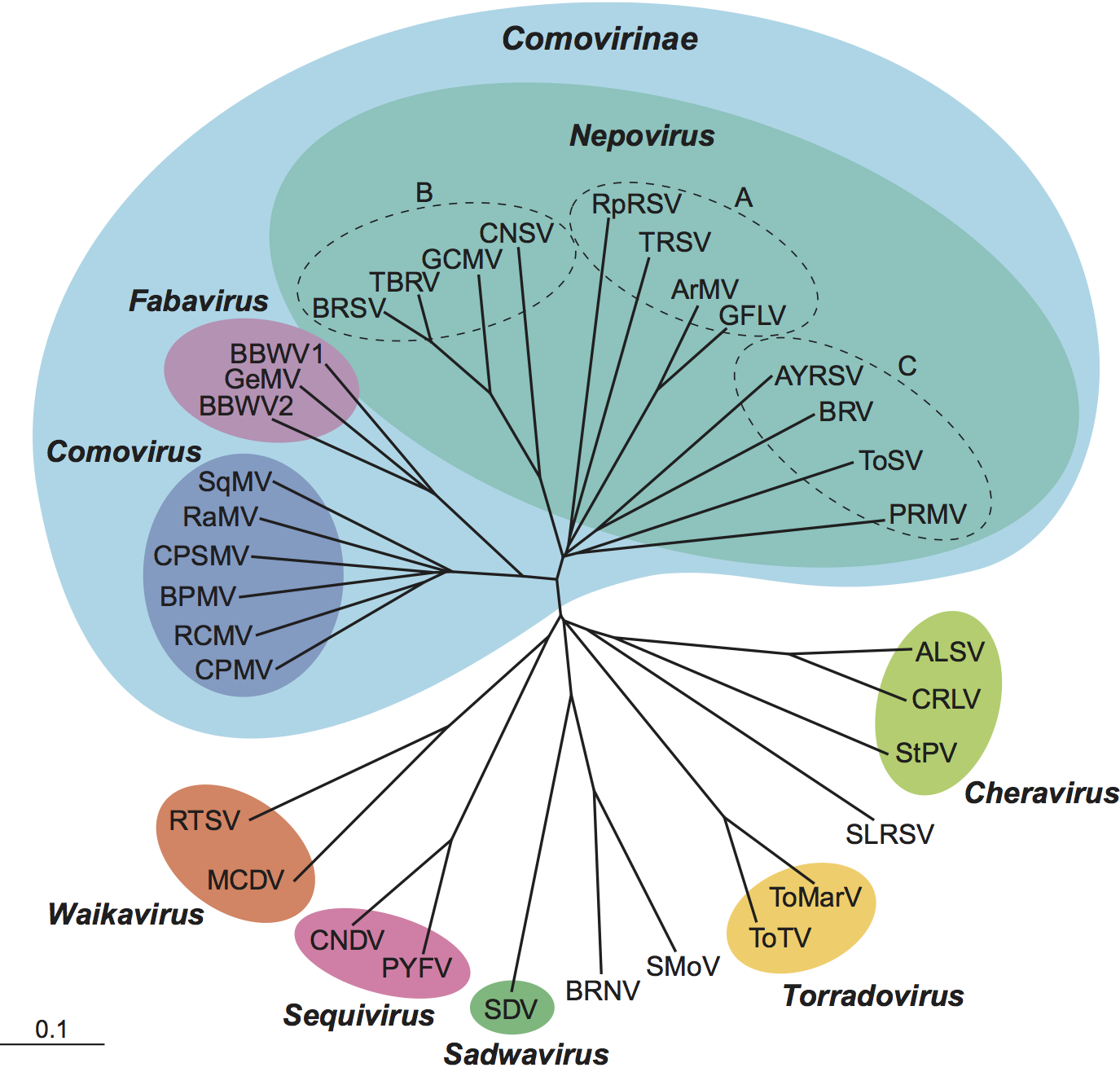
The conserved Pro-Pol region, delineated by the “CG” motif of the 3C-like proteinase and the “GDD” motif of the polymerase, has been used to determine the relationship among picornavirales. Comparison of the Pro-Pol sequence among members of the family Secoviridae allows the definition of branches that generally correspond to the distinct genera. Members of the subfamily Comovirinae (genera Comovirus, Fabavirus and Nepovirus) are more closely related to each other than to other genera within the family (Figure 5). Within this subfamily, fabaviruses and comoviruses are more closely related to each other than to nepoviruses. Nepovirus subgroups are not clearly separated in the Pro-Pol tree (with the exception of subgroup B which constitutes a separate branch) but are more clearly separated in phylogenetic trees using the CP sequence (not shown).
Similarity with other taxa
Secovirids are related to members of other families in the order Picornavirales. They all share a common virion structure, organization of the replication block within the polyproteins and conserved properties of the replication proteins, including the 3C-like proteinase. Secovirids are also related to members of the families Potyviridae and Caliciviridae in some aspects (common replication block, polyprotein strategy, VPg bound to the 5′ end of the genome and poly(A) tail at the 3′ end of the genome) but differ in other properties.
Derivation of names
Seco: derived from the amalgamation of the previous families Sequiviridae and Comoviridae.
Como: from cowpea mosaic virus, the type member.
Faba: derived from the Latin faba, “bean”; also Vicia faba, broad bean.
Nepo: from nematode-transmitted, polyhedral particles.
Chera: from cherry rasp leaf virus, the type member.
Sadwa: from satsuma dwarf virus, the type member.
Torrado: derived from tomato torrado virus, the type member. In Spanish, torrado means “toasted” to refer to the severe necrosis (burnt-like phenotype) observed in the disease induced by ToTV.
Sequi: from Latin sequi, “follow”, “accompany” (in reference to the dependent aphid transmission of parsnip yellow fleck virus).
Waika: from Japanese, describing the symptoms induced in rice by infection with rice tungro spherical virus alone (i.e. in the absence of rice tungro bacilliform virus).
Further reading
Choi, 2008 I. Choi, B.W.J. Mahy, M.H. Van Regenmortel, SequivirusesEncyclopedia of Virology. In: B.W.J. Mahy, M.H. Van Regenmortel, Encyclopedia of Virology. Elsevier, Oxford2008546–551.
Iwanami, 2008 T. Iwanami, B.W.J. Mahy, M.H. Van Regenmortel, SadwavirusEncyclopedia of Virology. In: B.W.J. Mahy, M.H. Van Regenmortel, Encyclopedia of Virology. Elsevier, Oxford2008523–526.
Le Gall et al., 2007 O. Le Gall, H. Sanfacon, M. Ikegami, T. Iwanami, T. Jones, A. Karasev, K. Lehto, J. Wellink, T. Wetzel, N. Yoshikawa, Cheravirus and Sadwavirus: two unassigned genera of plant positive-sense single-stranded RNA viruses formerly considered atypical members of the genus Nepovirus (family Comoviridae). Arch. Virol. 159 (2007) 1767–1774.
Lin and Johnson, 2003 T. Lin, J.E. Johnson, Structures of picorna-like plant viruses: implications and applications. Adv. Virus Res. 62 (2003) 167–239.
Lomonossoff, 2008 G. Lomonossoff, B.W.J. Mahy, M.H. Van Regenmortel, Cowpea mosaic virusEncyclopedia of Virology. In: B.W.J. Mahy, M.H. Van Regenmortel, Encyclopedia of Virology. Elsevier, Oxford2008.
Pouwels et al., 2002 J. Pouwels, J.E. Carette, J. Van Lent, J. Wellink, Cowpea mosaic virus: effects on host cell processes. Mol. Plant Pathol. 3 (2002) 411–418.
Sanfacon, 2008 H. Sanfacon, B.W.J. Mahy, M.H. Van Regenmortel, NepovirusEncyclopedia of Virology. In: B.W.J. Mahy, M.H. Van Regenmortel, Encyclopedia of Virology. Elsevier, Oxford2008405–413.
Sanfacon, 2009 H. Sanfacon, Secoviridae: The amalgamation of the families Sequiviridae and ComoviridaeEncyclopedia of Life Sciences. John Wiley & Sons, Chichester2009.
Sanfacon et al., 2009 H. Sanfacon, J. Wellink, O. Le Gall, A. Karasev, R. van der Vlugt, T. Wetzel, Secoviridae: a proposed family of plant viruses within the order Picornavirales that combines the families Sequiviridae and Comoviridae, the unassigned genera Cheravirus and Sadwavirus, and the proposed genus. Torradovirus. Arch. Virol. 154 (2009) 899–907.
Susi, 2004 P. Susi, Black currant reversion virus, a mite-transmitted nepovirus. Mol. Plant Pathol. 5 (2004) 167–173.
Contributed by
Sanfaçon, H., Iwanami, T., Karasev, A.V., van der Vlugt, R., Wellink, J., Wetzel, T. and Yoshikawa, N.
Figures
Figure 1 (Top left): Molecular rendering of the cowpea mosaic virus particle (Lin et al. (1999). Virology265, 20-34; with permission). (Top central): Diagrammatic representation of a T=1 lattice. A=Small capsid protein, B=C-terminal domain of the large capsid protein and C=N-terminal domain of the large capsid protein. (Top right): Molecular rendering of the red clover mottle virus particle (Lin et al. 2000, with permission). (Center): Diagram of the three types of comovirus particles with the B-particle containing one molecule of RNA-1, the M-particle containing one molecule of RNA-2 and the T-particle being empty. (Bottom): Negative contrast electron micrograph of particles of cowpea mosaic virus. The bar represents 100 nm.
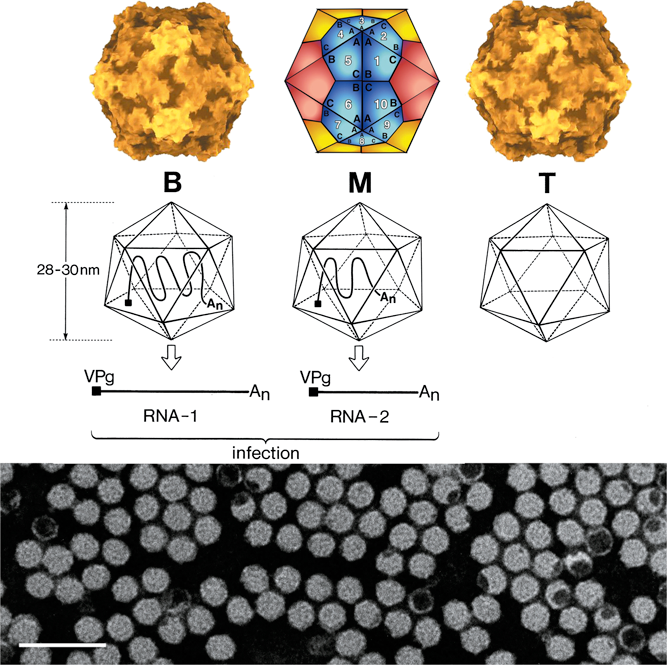
Figure 2 Architecture of the capsid of members of the family Secoviridae. In sequiviruses, waikaviruses, cheraviruses and torradoviruses, each subunit is composed of three separate small coat proteins (CPs) each containing a single beta-barrel domain (VP1-VP3, top). In comoviruses, fabaviruses and sadwaviruses, the three beta-barrels are present in two CPs (VPL with two barrels and VPS with a single barrel, middle). In nepoviruses, the single CP is folded in three barrels (bottom).
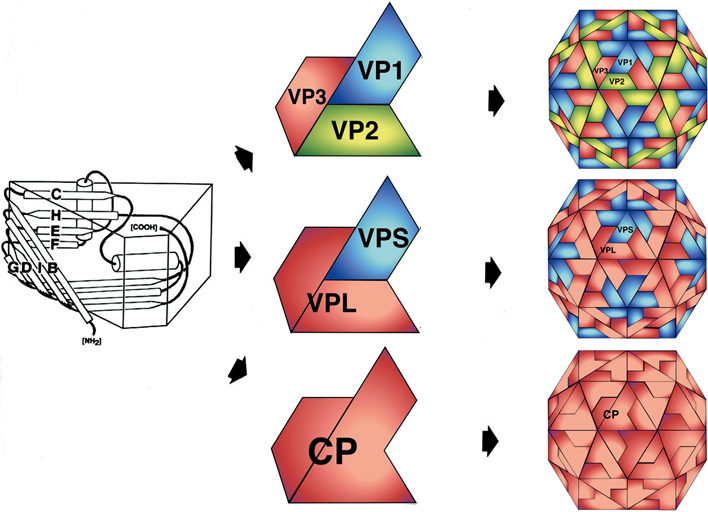
Figure 3 Genome organization of representative members of the family Secoviridae. Each RNA is shown with the ORFs represented with the boxes. Circles depict VPg molecules covalently attached at the 5 end of the RNAs. Black circles represent VPg confirmed experimentally and open circles represent putative VPgs. Poly(A) tails are represented at the 3 end of the RNAs when present [A(n)]. Red arrows above the sequences represent regions of extensive sequence identity between RNAs 1 and 2. Protein domains with conserved motifs for the putative NTP-binding protein (NTB, shown in orange), VPg (purple), proteinase (Pro, yellow), RNA-dependent RNA polymerase (Pol, red), movement protein (MP, green) and coat protein(s) (CP, blue) are shown. The star represents a conserved motif found in the Co-Pro protein of comoviruses and in the equivalent protein of other viruses. Proteinase cleavage sites identified experimentally or deduced by sequence comparisons are shown by the solid or dotted vertical lines, respectively. Possible ORFs in the genome of waikaviruses are shown with the dotted squares and putative subgenomic RNAs are shown by dotted arrows below the waikavirus genome. For each virus, the genomic organization is shown for the type isolate as described in the text.
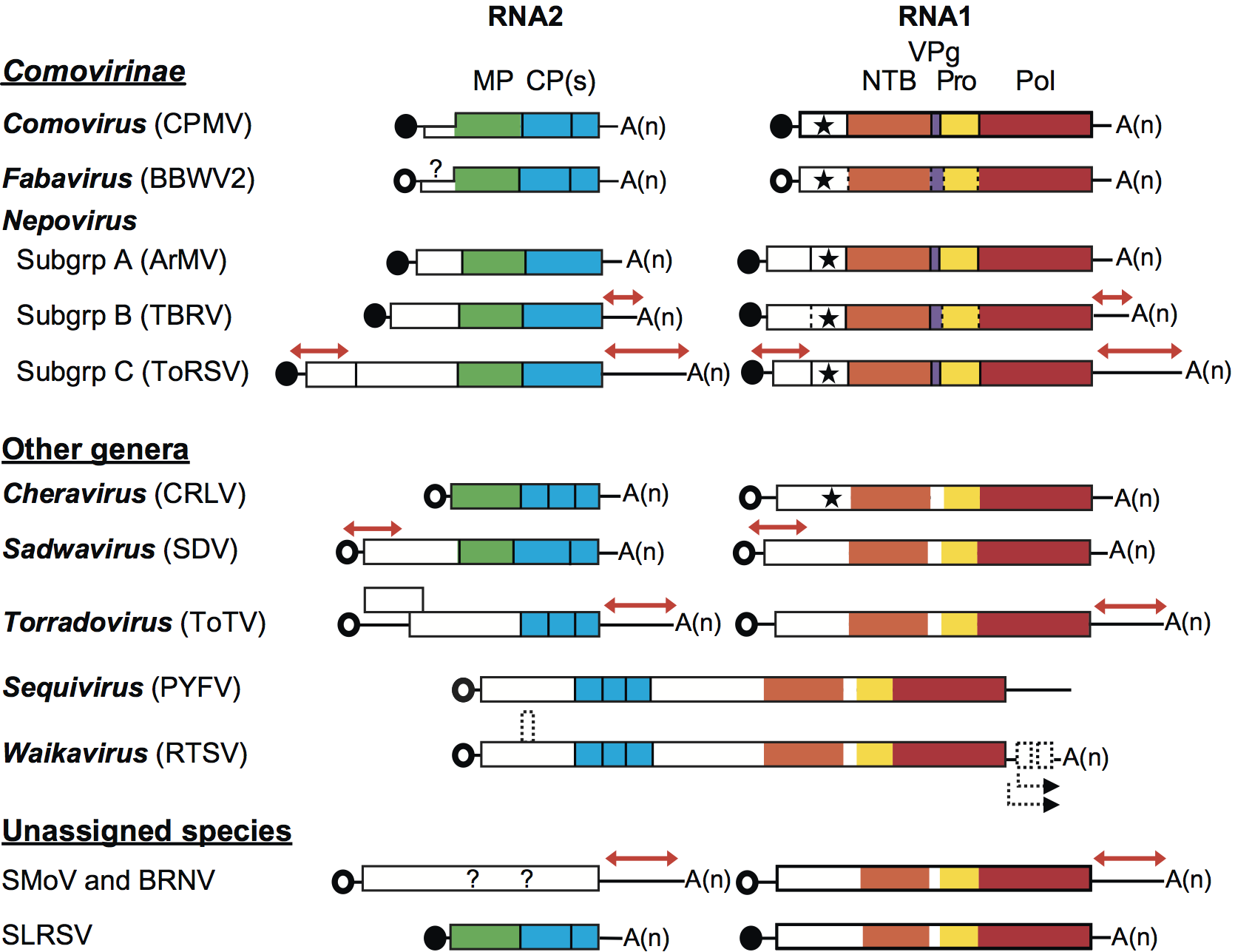
Figure 4 Genome organization and polyprotein processing of cowpea mosaic virus. The ORFs are boxed and the function of the proteins is indicated. MP: movement protein; CPL and CPS: large and small coat proteins; Co-Pro: proteinase co-factor; NTB: NTP-binding proteins; Pro: proteinase; Pol: RNA-dependent RNA polymerase. Proteolytic cleavage sites are indicated on the polyproteins with the vertical lines. All intermediate and final cleavage products have been detected in infected cells. The black circles at the 5 end of the RNA represents the VPg, and A(n) at the 3 end the poly-A tail.
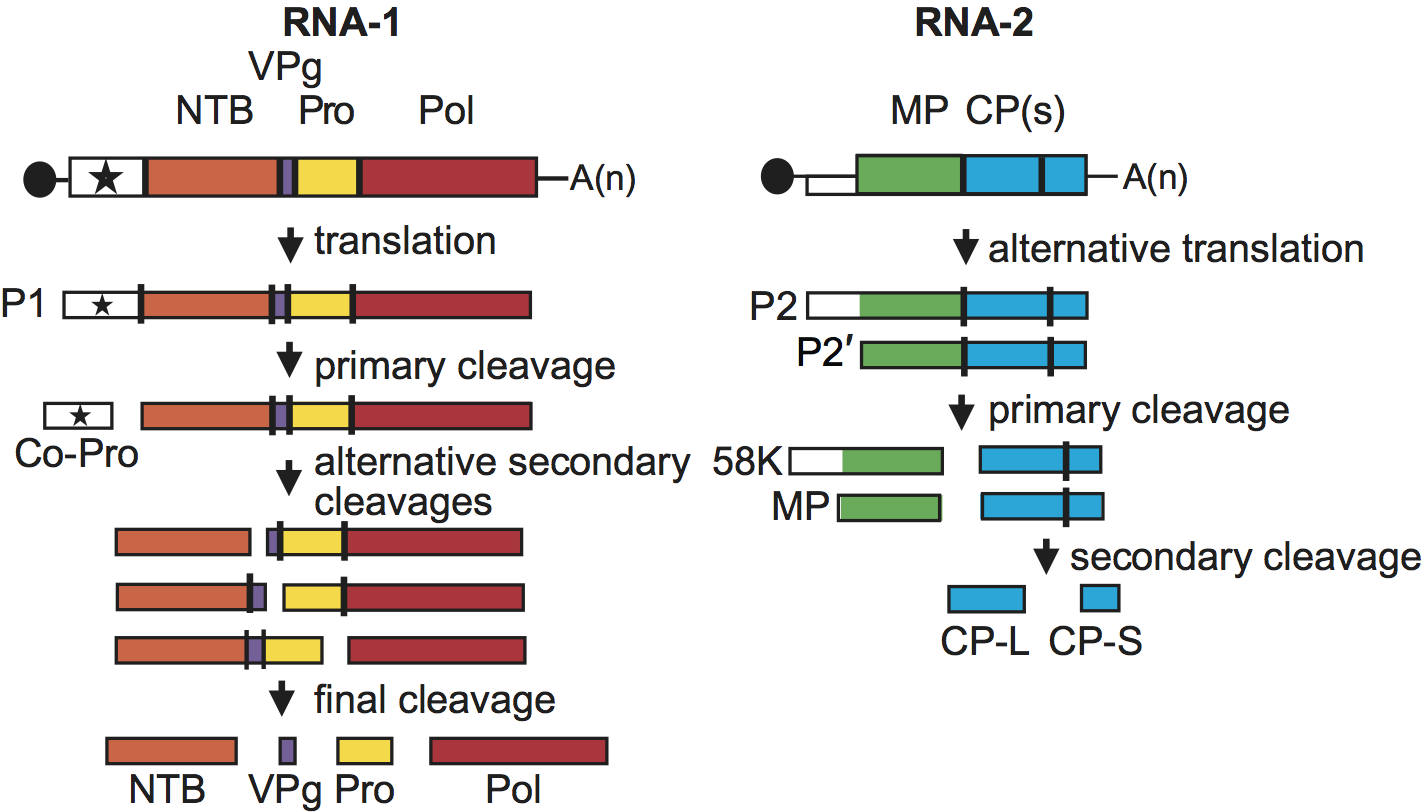
Figure 5 Hierarchical clustering of members of the family Secoviridae based on the amino acid sequences of the conserved domains between the CG motif of the 3C-proteinase and the GDD motif of the polymerase (Pro-Pol region). Results are presented as an unrooted radial tree. The bar represents a p-distance of 0.1. Different genera within the family are shown with the color ovals. The subfamily Comovirinae is shown with the light blue shading. Unfilled circles with the letters represent the different nepovirus subgroups. For each species, the sequence of the type isolate was used for the alignments (see species tables for the sequence accession numbers).