Order: Picornavirales
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Introduction
The order Picornavirales contains viruses with a monopartitite or bipartite positive-strand RNA genome that share the following properties: auto-proteolytically processed polyprotein(s), a common three-domain replication block (Hel-Pro-Pol domain consisting of a superfamily III helicase, a proteinase with a chymotrypsin-like structure and a superfamily I RNA-dependent RNA polymerase) and non-enveloped icosahedral virions approximately 30 nm in diameter with a pseudo-T=3 symmetry. The RNAs are usually characterized by the presence of a small VPg protein (typical 3–4 kDa) linked to their 5′ end and a poly(A) tail at their 3′ end. Members of the family Picornaviridae (genus Enterovirus) and of the family Secoviridae (genus Comovirus) were the first characterized members of the order and infect vertebrates and plants, respectively. The order also includes viruses infecting invertebrates (families Dicistroviridae and Iflaviridae) or algae (family Marnaviridae). Large-scale environmental genomic studies suggest the presence of a large number of uncharacterized picorna-like viruses in the ocean.
Virion properties
Morphology
Virions are non-enveloped icosahedral particles approximately 30 nm in diameter with a pseudo-T=3 symmetry. Virus preparations often contain empty virus particles.
Physicochemical and physical properties
Empty virus particles sediment with S20,W values between 50 and 80S. Viruses with a single large genomic RNA (families Picornaviridae, Iflaviridae and Dicistroviridae and genera Sequivirus and Waikavirus) sediment with S20,W values of 140–190S. Viruses with a bipartite genome (family Secoviridae with the exception of the genera Sequivirus and Waikavirus) encapsidate each RNA separately. Virions containing RNA1 sediment at 110–135S and virions containing RNA2 sediment at 84–128S.
Nucleic acid
Most members of the order have a single molecule of positive sense RNA ranging between 7,000 and 12,500 nt in length. Some members of the order (genera Comovirus, Fabavirus, Nepovirus, Sadwavirus, Cheravirus and Torradovirus all in the family Secoviridae) have a bipartite genome with an RNA1 ranging in size between 5800 and 8400 nt and an RNA2 between 3200 and 7300 nts. The presence of a small (3–5 kDa) VPg linked to the 5′ end of the RNAs has been confirmed for most members of the order; comparative genomics strongly suggests that this property is universally conserved. Viral RNAs are normally polyadenylated at their 3′ end. The only known exception is the genomic RNA of parsnip yellow fleck virus (genus Sequivirus, family Secoviridae) which is apparently not polyadenylated.
Proteins
The capsid contains 60 units, each consisting of three paralogous jelly-roll domains. Each jelly-roll domain is unique in sequence but folds in a similar 8-stranded beta-barrel structure. In most members of the order, the three jelly-roll domains are contained in three separate coat proteins (CP) of approximately 25 kDa each. However, in some members of the family Secoviridae, the three jelly-roll domains can be contained in a single large CP of approximately 60 kDa (genus Nepovirus) or in two CPs, a 40–45 kDa CP containing two jelly-roll domains and a 21–29 kDa CP containing one jelly-roll domain (genera Comovirus, Fabavirus and Sadwavirus). An additional unrelated small structural protein, termed VP4, is encoded by the genome of some members of the families Picornaviridae (upstream of VP2), Dicistroviridae and Iflaviridae (upstream of VP3). This small protein is encapsidated within the virion.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The viral RNA is infectious and serves as a template for replication and as mRNA. In the case of viruses with a bipartite genome, RNA1 can replicate independently of RNA2 within the first infected cell but RNA2 is required for encapsidation and for cell-to-cell movement. The combined size of the 5′ and 3′ UTRs is <15% of the genome. They contain sequence motifs and secondary structures necessary for replication/translation. The 5′-UTR is larger than 3′-UTR in animal viruses. The RNAs of viruses in the family Secoviridae often contain a much larger 3′-UTR (Figure 1).
Figure 1 Genome organization of representative members of the order Picornavirales. Each RNA is shown with the ORF(s) represented with the boxes. Circles depict VPg molecules covalently attached at the 5′ end of the RNAs. Black circles represent VPg confirmed experimentally and open circles represent putative VPgs. Poly(A) tails are represented at the 3′ end of the RNAs [A(n)]. Conserved motifs for the helicase (orange diamonds), 3C or 3C-like proteinase (yellow diamonds), RNA-dependent RNA polymerase (red diamonds) and the domains for coat protein(s) (blue rectangles) are shown. Please refer to chapters describing each family for further details (presence and function of additional protein domains, position of cleavage sites, etc.). When characterized, internal ribosome entry sites are shown with the black stars (families Picornaviridae and Dicistroviridae). This has not been studied for other members of the order. Abbreviation for virus species are as follows: human enterovirus C (HEV-C), Theiler’s murine encephyalomyocarditis virus (TMEV), foot-and-mouth disease virus (FMDV), infectious flacherie virus (IFV), cricket paralysis virus (CrPV), Heterosigma akashiwo RNA virus (HaRNAV), rice tungro spherical virus (RTSV), cowpea mosaic virus (CPMV), tomato ringspot virus (ToRSV), tomato torrado virus (ToTV).
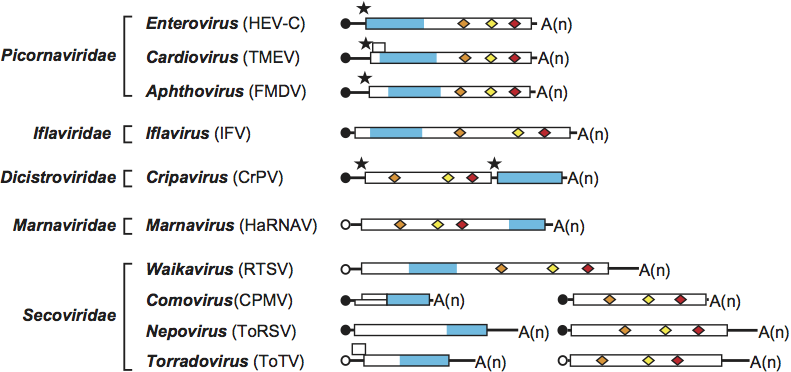
The RNA genomes of viruses in the order are generally monocistronic with a single ORF that encodes a single large polyprotein. For many members of the order, translation of the unique ORF has been shown to be directed by an internal ribosome entry site (IRES) located in the 5′ UTR. In viruses with a monopartite genome, the replication proteins (Hel, Pro, Pol) are generally present in the C-terminal region of the unique polyprotein and structural proteins are present in the N-terminal region of the polyprotein (family Picornaviridae and Iflaviridae, genera Sequivirus and Waikavirus). However, in the families Dicistroviridae and Marnaviridae, the structural proteins are located downstream of the replication proteins either within a single polyprotein (Marnaviridae) or as a separate polyprotein (Dicistroviridae). In bipartite members of the order (genera Comovirus, Fabavirus, Nepovirus, Sadwavirus, Cheravirus and Torradovirus), the RNA1-encoded polyprotein contains the domains for replication proteins and the RNA2-encoded polyprotein includes the domains for structural proteins and the movement protein (located upstream of the structural proteins).
There are some exceptions to the general rule that one polyprotein is encoded by each viral RNA. In the family Dicistroviridae, the RNA genome is dicistronic with two non-overlapping ORFs that are separated by an intergenic untranslated region (IGR). The first ORF encodes the replication proteins and the second ORF codes for structural proteins. Translation of the second ORF is directed by an IRES located in the IGR. In the genus Comovirus, two polyproteins are synthesized from the RNA2 single long ORF. The second shorter polyprotein is produced by translation initiation at an internal AUG. In the genus Torradovirus, two overlapping ORFs are found on RNA2. The function of the putative protein encoded by the first ORF and the mode of translation of the two ORFs have not been investigated. In a virus of the genus Cardiovirus, a small ORF overlaps with the beginning of the major ORF.
Some viruses within the order possess multiple VPgs encoded in tandem (three in the species Foot-and-mouth disease virus, two to six copies in most members of the family Dicistroviridae). It has been suggested that the unclassified seal picornavirus 1 has two VPgs. The VPg ORF(s) lie(s) between the 2Chel and 3Cpro genome regions. In picornaviruses, VPg is covalently attached, via a tyrosine at position 3 to the 5′ uracil of the genome. In comoviruses it is the amino-terminal serine which is used.
Most of the polyprotein cleavage sites are processed by a cysteine proteinase with a chymotrypsin-like structure. This proteinase is termed 3C proteinase for members of the family Picornaviridae and 3C-like proteinase in other members of the order. In the case of Heterosigma akashiwo RNA virus (the only characterized member of the family Marnaviridae), the cysteine in the catalytic triad of the proteinase is replaced by a serine. 3C and 3C-like proteinases usually cut after a glutamine or glutamate residue, although there are notable exceptions in the families Secoviridae and Marnaviridae. In the case of viruses with a bipartite genome, the 3C-like proteinase is encoded by RNA1 and the proteinase cleaves in trans the RNA2-encoded polyprotein. Members of the genus Enterovirus (family Picornaviridae) encode an additional proteinase, 2A, that cleaves at its own N-terminus in cis. The leader protein of members of the genera Aphthovirus and Erbovirus (family Picornaviridae) also has proteolytic activity.
For several members of the families Picornaviridae and Secoviridae, replication has been shown to occur in the cytoplasm in association with intracellular membranes. Several viral proteins interact directly with these membranes and anchor the replication complex to the membranes. Mode of replication of other members of the order has not been investigated.
Antigenic properties
These vary depending on the genus.
Biological properties
Members of the order infect a wide range of hosts including vertebrates (family Picornaviridae), plants (family Secoviridae), arthropods (family Dicistroviridae and Iflaviridae) and unicellular organisms (family Marnaviridae). Specific host range and transmission properties differ among individual genera.
Phylogenetic relationships within the order
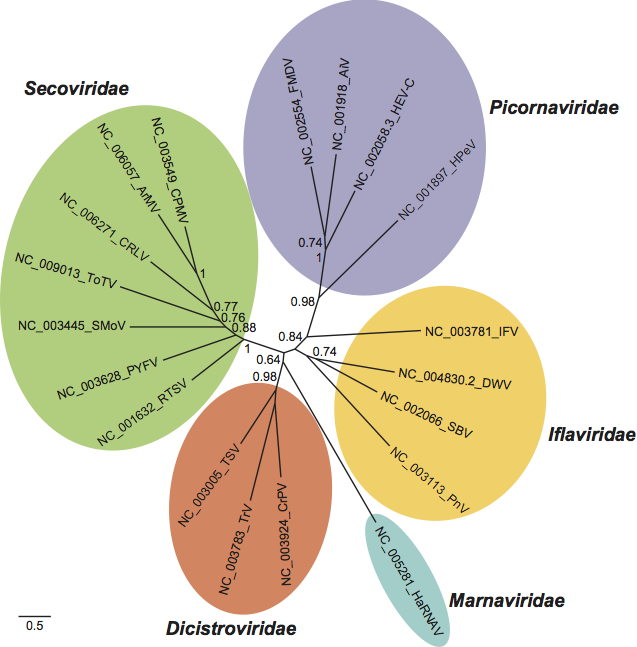
In an unrooted maximum-likelihood tree built for representatives of the order using the amino acid sequence contained in the proteinase-polymerase region, all families except the Iflaviridae form monophyletic branches (Figure 2).
Similarity with other taxa
Presence of the Hel-Pro-Pol domain within polyproteins is a property that is shared with members of the families Caliciviridae, Potyviridae and some unclassified viruses. However, members of these families differ in several properties including virus particle structure (elongated particles with helical symmetry for potyvirids and icosahedral particles but with a true T=3 structure for calicivirids), type of helicase (superfamily II helicase for potyvirids) and size of VPg (25 kDa for potyvirids and 12–15 kDa for calicivirids). In addition, some members of the family Caliciviridae express their structural proteins from a subgenomic RNA. Members of the order Picornavirales are not known to produce subgenomic RNAs. There are also other virus families which use either jelly-roll-based capsids and/or 3C-like proteases.
Derivation of name
The name is derived from the family Picornaviridae.
Further reading
Le Gall et al., 2008 O. Le Gall, P. Christian, C.M. Fauquet, A.M.Q. King, N.J. Knowles, N. Nakashima, G. Stanway, A.E. Gorbalenya, Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch. Virol. 153 (2008) 715–727.
Contributed by
Sanfaçon, H., Gorbalenya, A.E., Knowles, N.J. and Chen, Y.P.
