Family: Picornaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
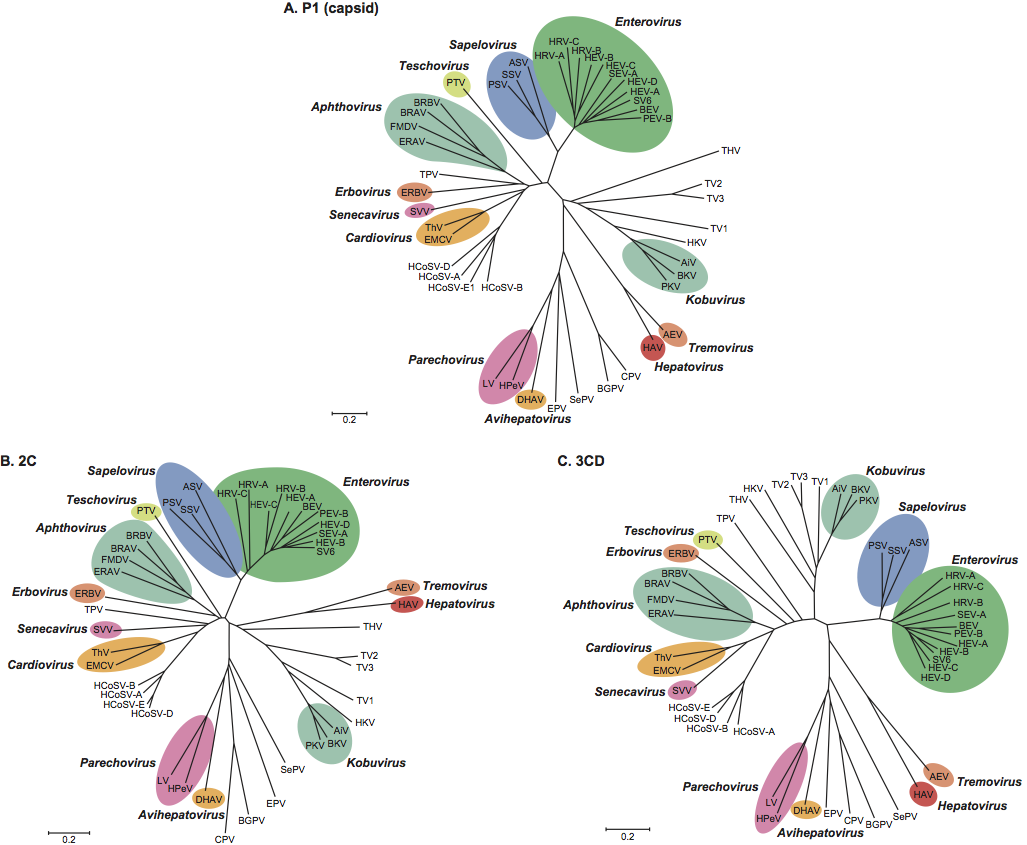
Virions consist of a capsid, with no envelope, surrounding a core of ssRNA. Hydrated native particles are 30 nm in diameter, but vary from 22 to 30 nm in electron micrographs due to drying and flattening during preparation. Electron micrographs reveal no projections on most picornaviruses, the virion appearing as an almost featureless sphere; however, kobuviruses, and possibly parechoviruses, show a surface structure that is distinct from small round structured viruses (astroviruses and caliciviruses) (Figure 1). The capsid is composed of 60 identical units (protomers), each consisting of three surface proteins, 1B, 1C and 1D, of 24–41 kDa, and, in most picornaviruses, an internal protein, 1A of 5.5–13.5 kDa; however, in some viruses 1AB (VP0) remains uncleaved. Total protomer is 80–97 kDa. Proteins 1A, 1B, 1C and 1D are also commonly named VP4, VP2, VP3 and VP1, respectively. Proteins 1B, 1C and 1D each possess a core structure comprising an eight-stranded β-sandwich (“β-barrel”). The β-barrels pack together in the capsid with T=l, pseudo T=3, icosahedral symmetry. These structural features are shared by the other members of the order Picornavirales. Genera differ in the external loops that interconnect the β strands. These loops account for differences in surface relief of each genus (Figure 1) and in thickness of the capsid wall. Assembly occurs via pentameric intermediates (pentamer=five protomers). Proteins within each pentamer are held together by an internal network formed from the N-termini of the three major capsid proteins (CPs), the C-termini lying on the outer capsid surface. Empty capsids, which are produced by some picornaviruses, are very similar to virions, except that 1A and 1B are normally replaced by the uncleaved precursor, 1AB.
Physicochemical and physical properties
Virion Mr is 8×106 to 9×106, S20,w is 140–165S (empty particle S20,w is 70–80S). Buoyant density in CsCl is 1.33–1.45 g cm−3, depending on the genus. Some species are unstable below pH 7; many are less stable at low ionic strength than at high ionic strength. Virions are insensitive to ether, chloroform, or non-ionic detergents. Viruses are inactivated by light when grown with, or in the presence of photodynamic dyes such as neutral red or proflavin. Thermal stability varies with viruses as does stabilization by divalent cations.
Nucleic acid
Virions contain one molecule of positive sense, ssRNA, 7–8.8 kb in size, and possessing a single long ORF. A poly(A) tail, heterogeneous in length, is located after the 3′-terminal heteropolymeric sequence. A small protein, VPg (ca. 2.2–3.9 kDa), is linked covalently to the 5′ terminus. The UTRs at both termini contain regions of secondary structure which are essential to genome function. The long 5′-UTR (0.5–1.5 kb) includes a 5′-terminal domain involved in replication (e.g. the poliovirus “clover-leaf”) and an IRES of 220–450 nt upstream of the translational start site; most picornaviral IRES elements can be assigned to one of four types (I to IV), according to their secondary structure. Between the 5′-terminal domain and the IRES there may be one, or more, pseudoknots and/or a poly(C) tract (Figure 2). The 3′-UTR, which may also contain a pseudoknot, ranges from 40 to 330 nt in length. The overall sequence identity between the genomes of viruses of different genera is typically less than 40%. The G+C content of picornavirus genomes ranges from 35 to almost 60%.
Proteins
In addition to the major CPs, 1A, 1B, 1C and 1D, and 3B (VPg), described above, small amounts of 1AB (VP0) are commonly seen in lieu of one or more copies of 1A and 1B. Protein 1A is small in hepatoviruses, and 1AB is uncleaved in avihepatoviruses, kobuviruses, parechoviruses and a number of unclassified picornaviruses. Traces of other proteins, including the viral RdRp, 3Dpol, may also be present in purified virus preparations.
Lipids
Some picornaviruses carry a sphingosine-like molecule (“pocket factor”) in a cavity (“pocket”) located inside 1D. Protein 1A, where present, generally has a molecule of myristic acid covalently attached to the amino terminal glycine.
Carbohydrates
None of the viral proteins is glycosylated.
Genome organization and replication
The virion RNA is infectious and serves as both the genome and the viral mRNA. Gene maps are shown in Figure 3. Initiation of protein synthesis is stimulated by the IRES. Translation of the single ORF produces the polyprotein precursor 240–250 kDa to the structural proteins (derived from the P1 region of the genome) and the non-structural proteins (from the P2 and P3 regions). In some viruses P1 is preceded by a leader protein (L). The polyprotein is cleaved to functional proteins by specific proteases contained within it. Intermediates are denoted by letter combinations (e.g. 3CD, the uncleaved precursor of 3C and 3D). The viral proteases are as follows: 3Cpro, a chymotrypsin-like cysteine protease encoded by all picornaviruses, performs most of the cleavages. In enteroviruses, and possibly sapeloviruses, 2A is also associated with proteolytic activity (2Apro); the 2A of aphthoviruses, avihepatoviruses, cardioviruses, erboviruses, senecaviruses, teschoviruses and Ljungan virus (genus Parechovirus) contains a NPG↓P motif (↓=cleavage site) and acts only in cis. The leader proteins of aphthoviruses and erboviruses have proteolytic activity (Lpro). Some intermediates are stable and serve functions distinct from those of their cleavage products (e.g. cleavage of poliovirus P1 by 3CDpro, not by 3Cpro). Where it occurs, the cleavage of 1AB, which accompanies RNA encapsidation, is thought to be autocatalytic, but the precise mechanism is unknown.
A typical picornavirus genome layout may be represented by the following:
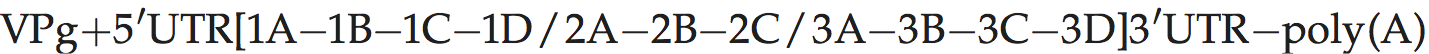
Where “[” and “]” define the extent of the polyprotein-coding region, “/” represents primary cleavages and “−” represents the final cleavages. Where a particular polypeptide is present only in some members of the genus it can be shown between parentheses. This schema can also be used to indicate some protein functions or amino acid motifs where they differ between viruses (e.g. 2Apro or 2Anpgp or 2AH-box/NC). There may be multiple copies of a particular genomic regions in the picornavirus genome, including repeated copies of one particular region (e.g. three 3Bs in the FMDV genome) or different types of a particular region (e.g. two different 2A motifs in Ljungan virus of the genus Parechovirus and three different 2A motifs in the duck hepatitis A virus genome of the genus Avihepatovirus).
Replication of viral RNA occurs in complexes associated with cytoplasmic membranes. These complexes contain proteins derived from the whole of the 2BC-P3 region of the polyprotein, including the polymerase (3Dpol, an RNA chain-elongating enzyme), and 2C (an ATPase containing a nucleotide binding sequence motif). The poliovirus 3Cpro component has been shown to be required for binding to the 5′-terminal RNA cloverleaf. The short virus-encoded protein, VPg, acts as a transcription primer for both positive and negative strand RNA synthesis. Prior to transcription two uridine residues are covalently linked to the conserved tyrosine at position 3 in VPg to form VPgpUpUOH via a templating mechanism involving a cis-acting replication element (cre) and the virus 3D polymerase. The cre is a stem loop containing the sequence “AAAC” in the loop and is found at various places in the genome depending on virus species/genus. Many compounds that specifically inhibit replication have been described. Mutants resistant to, or dependent on, drugs have been reported. Genetic recombination, complementation, and phenotypic mixing occur. Defective particles, carrying deletions in the CPs or L, have been produced experimentally but have not been observed in natural virus populations.
Antigenic properties
Serotypes are classified, depending on genus, by cross-protection, neutralization of infectivity, complement-fixation, specific ELISA using a capture format or immunodiffusion. Some serotypes can be identified using hemagglutination-inhibition. Antigenic sites, defined by mutations that confer resistance to neutralization by monoclonal antibodies, typically number 3 or 4 per protomer.
Biological properties
Most picornaviruses are specific for one, or a very few host species [exceptions are foot-and-mouth disease virus (FMDV) and encephalomyocarditis virus (EMCV)]. Members of most species can be grown in cell culture. Resistant host cells (e.g., mouse cells in the case of the primate-specific polioviruses) can often be infected (for a single round) by transfection with naked, infectious RNA. Transmission is horizontal, mainly by fecal–oral, fomite or airborne routes. Transmission by arthropod vectors is not known, although EMCV has been isolated from mosquitoes and ticks and poliovirus from flies; therefore mechanical transmission may be possible.
Infection is generally cytolytic, but persistent infections are common with some species and reported with others. Poliovirus infected cells undergo extensive vacuolation as membranes are reorganized into viral replication complexes. Infection may be accompanied by rapid inhibition of cap-dependent translation of cellular mRNAs (2Apro of poliovirus and Lpro of aphthovirus are each powerful inhibitors), mRNA synthesis, and the cellular secretory pathway (poliovirus 2B and 3A have been implicated).
Species demarcation criteria in the family
A picornavirus species is a class of phylogenetically-related serotypes or strains which would normally be expected to share (i) a limited range of hosts and cellular receptors, (ii) a significant degree of compatibility in proteolytic processing, replication, encapsidation and genetic recombination, and (iii) essentially identical genome maps. The polyprotein sequences of viruses in different genera differ by at least 58% aa identity. To distinguish virus names from species names where they have been the same the Picornaviridae Study Group recommends that viruses be assigned a type number, e.g. encephalomyocarditis virus will now be known as encephalomyocarditis virus 1 (EMCV-1); this currently affects some virus names in the following genera: Cardiovirus, Aphthovirus, Hepatovirus, Kobuvirus, Sapelovirus, Senecavirus and Tremovirus.
Genus Enterovirus
Type species Human enterovirus C
Virion properties
Morphology
CPs 1B, 1C and 1D of the human enteroviruses and rhinoviruses are among the largest in the family (VP1-3 chain lengths, 238–302 aa), and this is reflected in the typically long inter-β-strand loops, the larger than average thickness of the capsid wall (46 Å), and a surface relief that is strongly pronounced compared to most other picornaviruses. Encircling a raised area at the five-fold axis is a 25 Å deep groove, or “canyon”, into which the cellular receptor for poliovirus binds. The binding site for the pocket factor lies beneath the floor of this canyon within the 1D β-barrel. Virions can be converted by a variety of treatments (gentle heating, binding to receptor, or some neutralizing antibodies) to altered (“A”) particles of 135S which lack 1A (VP4) and possess altered antigenicity.
Physicochemical and physical properties
Acid stability is variable. The virions of most enterovirus species are stable at pH 3.0, while those of the rhinovirus species are unstable below pH 5–6. Similarly, the buoyant density in CsCl of the enterovirus virions is 1.30–1.34 g cm−3, while the rhinoviruses range from 1.38 to 1.42 g cm−3. Sometimes a small proportion (about 1% of the population) of heavy particles (density: 1.43 g cm−3) can be observed in the enteroviruses. Empty capsids are often observed in virus preparations.
Nucleic acid
The genome contains a type I IRES and no poly(C) tract. The cre is located in 2C (HEV-A, HEV-B, HEV-C and HEV-D) or 2A (HRV-A) or 1D (HRV-B) or 1B (HEV-C). Sequence identities for different enteroviruses, or between enteroviruses and rhinoviruses, are more than 50% over the genome as a whole although it may be greater or less than this for particular genomic regions. The 5′-UTR of human rhinoviruses is shorter (ca. 650 nt) than that of enteroviruses, due to a deletion of approximately 100 nt between the IRES and the translation start site. Some members of HEV-C and HEV-D also have smaller deletions in this region. Bovine enteroviruses have a non-perfect duplication of the first ~100 nucleotides, allowing the formation of a second clover-leaf-like RNA structure. Porcine enteroviruses have an insertion of about 30 nt approximately 65 nt from the 5′ end of the genome resulting in a longer stem-loop D in the cloverleaf structure. Varying size deletions in the same region have been observed in some of the human enteroviruses.
Genome organization and replication
Genome layout:
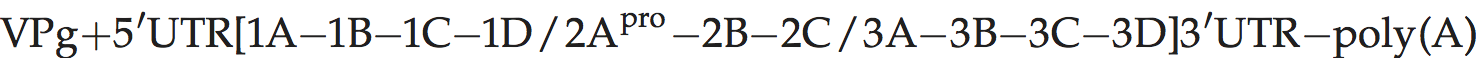
Genomes encode a single VPg and no L protein. Protease 2Apro, which is related to the family of small bacterial serine proteases, cleaves the polyprotein at its own N-terminus. Certain hydrophobic molecules that bind to the capsid in competition with pocket factor exert a powerful antiviral action by interfering with receptor binding and/or uncoating. Antiviral, pocket-binding drugs have been described.
Antigenic properties
Native virions are antigenically serotype-specific (designated “N” or “D” for poliovirus), whereas “A” particles exhibit group specificity (designated “H” or “C” for poliovirus).
Biological properties
Viruses multiply primarily in the gastrointestinal tract or the upper respiratory tract or sometimes both, but they can also multiply in other tissues, e.g., nerve, muscle, etc. Infection may frequently be asymptomatic. Clinical manifestations include common cold, mild meningitis, encephalitis, myelitis, myocarditis and conjunctivitis. Swine vesicular disease virus is a variant of coxsackievirus B5 and causes a vesicular disease in pigs clinically indistinguishable from foot-and-mouth disease (genus Aphthovirus). Cap-dependent translation of host mRNA is inhibited by 2Apro, which cleaves the host eukaryotic initiation factor 4G (eIF-4G). Many different cell surface molecules, many of them uncharacterized, serve as viral receptors. Well characterized receptor/virus interactions include poliovirus receptor (PVR) / polioviruses), coxsackievirus-adenovirus receptor (CAR)/coxsackie B viruses, intercellular adhesion molecule 1 (ICAM-l)/major-group rhinoviruses and some members of the Human enterovirus C species, low-density lipoprotein receptor (LDLR)/minor-group rhinoviruses, decay-accelerating factor (DAF)/various enteroviruses, integrin VLA-2/echovirus 1, and sialic acid/enterovirus D70.
Species demarcation criteria in the genus
Members of a species of the genus Enterovirus:
- share greater than 70% aa identity in the polyprotein
- share greater than 60% aa identity in P1
- share greater than 70% aa identity in the non-structural proteins 2C + 3CD
- share a limited range of host cell receptors
- share a limited natural host range
- have a genome base composition (G+C) which varies by no more than 2.5%
- share a significant degree of compatibility in proteolytic processing, replication, encapsidation and genetic recombination.
List of species in the genus Enterovirus
Certain viruses initially reported as novel echoviruses were later shown to have been misidentified. Thus E-8 is the same serotype as E-1, E-10 is now reovirus 1, E-28 is now human rhinovirus 1A, E-22 is now human parechovirus 1, E-23 is now human parechovirus 2. Similarly CV-A23 is the same serotype as E-9, and CV-A15 is the same serotype as CV-A11 and CV-A18 is the same as CV-A13. Hepatitis A virus 1 (HAV-1; genus Hepatovirus) was previously assigned the name enterovirus 72. Human rhinovirus 87 has been found to be a strain of EV-D68. A number of simian viruses (SV), previously listed as tentative members of the genus, have been moved to the genus Sapelovirus, species Simian sapelovirus and renamed simian sapelovirus (SSV) 1 (SV2), SSV-2 (SV 49) and SSV-3 (SV16, SV-18, SV42, SV44 and SV45). Simian agent 4 (SA4), SV4, SV28 and A2-plaque virus have been assigned to the species Simian enterovirus A. Simian enteroviruses N125 and N203 have been placed as a new type, EV-108, which is not yet assigned to a species. Similarly EV-103, SV6 and SV-47 also remain types unassigned to a species. Porcine enteroviruses (PEV) belonging to CPE group I (types 1–7 and 11–13) have been moved to the genus Teschovirus and renamed porcine teschovirus (PTV) 1–10. The species Porcine enterovirus A (PEV type 8; CPE group II) has been moved to the genus Sapelovirus and renamed Porcine sapelovirus (porcine sapelovirus 1).
|
Bovine enterovirus
|
|
|
|
|
|
Bovine enterovirus 1 [VG(5)27]
|
[D00214]
|
(BEV-1)
|
|
|
Bovine enterovirus 2 [BEV-261]
|
[DQ092770]
|
(BEV-2)
|
|
Human enterovirus A
|
|
|
|
|
|
Coxsackievirus A2 [Fleetwood]
|
[AY421760]
|
(CV-A2)
|
|
|
Coxsackievirus A3 [Olson]
|
[AY421761]
|
(CV-A3)
|
|
|
Coxsackievirus A4 [High Point]
|
[AY421762]
|
(CV-A4)
|
|
|
Coxsackievirus A5 [Swartz]
|
[AY421763]
|
(CV-A5)
|
|
|
Coxsackievirus A6 [Gdula]
|
[AY421764]
|
(CV-A6)
|
|
|
Coxsackievirus A7 [Parker]
|
[AY421765]
|
(CV-A7)
|
|
|
Coxsackievirus A8 [Donovan]
|
[AY421766]
|
(CV-A8)
|
|
|
Coxsackievirus A10 [Kowalik]
|
[AY421767]
|
(CV-A10)
|
|
|
Coxsackievirus A12 [Texas-12]
|
[AY421768]
|
(CV-A12)
|
|
|
Coxsackievirus A14 [G-14]
|
[AY421769]
|
(CV-A14)
|
|
|
Coxsackievirus A16 [G-10]
|
[U05876]
|
(CV-A16)
|
|
|
Enterovirus A71 [BrCr]
|
[U22521]
|
(EV-A71)
|
|
|
Enterovirus A76 [FRA91-10369]
|
[AY697458]
|
(EV-A76)
|
|
|
Enterovirus A89 [BAN00-10359]
|
[AY697459]
|
(EV-A89)
|
|
|
Enterovirus A90 [BAN99-10399]
|
[AY697460]
|
(EV-A90)
|
|
|
Enterovirus A91 [BAN00-10406]
|
[AY697461]
|
(EV-A91)
|
|
|
Enterovirus A92 [USA/GA99-RJg-7]
|
[EF667344]
|
(EV-A92)
|
|
|
Enterovirus A114 [BAN-11610]
|
|
(EV-A114)
|
|
|
Simian virus 19 [M19s]
|
[AF326754]
|
(SV19)
|
|
|
Simian virus 43 [OM112t]
|
[AF326761]
|
(SV43)
|
|
|
Simian virus 46 [OM22]
|
[AF326764]
|
(SV46)
|
|
|
baboon enterovirus A13 [A13]
|
[AF326750]
|
(BA13)
|
|
Human enterovirus B
|
|
|
|
|
|
Coxsackievirus B1 [Conn-5]
|
[AJ295196]
|
(CV-B1)
|
|
|
Coxsackievirus B2 [Ohio-1]
|
[AF081485]
|
(CV-B2)
|
|
|
Coxsackievirus B3 [Nancy]
|
[M88483]
|
(CV-B3)
|
|
|
Coxsackievirus B4 [JVB (Benshoeten)]
|
[X05690]
|
(CV-B4)
|
|
|
Coxsackievirus B5 [Faulkner]
|
[AF114383]
|
(CV-B5)
|
|
|
(including swine vesicular disease virus [UKG/27/72])
|
[D00435]
|
(SVDV)
|
|
|
Coxsackievirus B6 [Schmitt]
|
[AF039205, AF105342, AF114384]
|
(CV-B6)
|
|
|
Coxsackievirus A9 [Bozek]
|
[AJ295200, AY679743]
|
(CV-A9)
|
|
|
Echovirus 1 [Farouk]
|
[AF029859]
|
(E-1)
|
|
|
Echovirus 2 [Cornelis]
|
[AF465518, AY302545]
|
(E-2)
|
|
|
Echovirus 3 [Morrisey]
|
[AY302553]
|
(E-3)
|
|
|
Echovirus 4 [Pesascek]
|
[AY302557]
|
(E-4)
|
|
|
Echovirus 5 [Noyce]
|
[AF083069]
|
(E-5)
|
|
|
Echovirus 6 [D'Amori]
|
[AY302558]
|
(E-6)
|
|
|
Echovirus 7 [Wallace]
|
[AF465516, AY036579, AY302559]
|
(E-7)
|
|
|
Echovirus 9 [Hill]
|
[X84981]
|
(E-9)
|
|
|
Echovirus 11 [Gregory]
|
[X80059]
|
(E-11)
|
|
|
Echovirus 12 [Travis]
|
[X79047]
|
(E-12)
|
|
|
Echovirus 13 [Del Carmen]
|
[AY302539]
|
(E-13)
|
|
|
Echovirus 14 [Tow]
|
[AY302540]
|
(E-14)
|
|
|
Echovirus 15 [Ch 96-51]
|
[AY302541]
|
(E-15)
|
|
|
Echovirus 16 [Harrington]
|
[AY302542]
|
(E-16)
|
|
|
Echovirus 17 [CHHE-29]
|
[AY302543]
|
(E-17)
|
|
|
Echovirus 18 [Metcalf]
|
[AF317694]
|
(E-18)
|
|
|
Echovirus 19 [Burke]
|
[AY302544]
|
(E-19)
|
|
|
Echovirus 20 [JV-1]
|
[AY302546]
|
(E-20)
|
|
|
Echovirus 21 [Farina]
|
[AY302547]
|
(E-21)
|
|
|
Echovirus 24 [DeCamp]
|
[AY302548]
|
(E-24)
|
|
|
Echovirus 25 [JV-4]
|
[AY302549]
|
(E-25)
|
|
|
Echovirus 26 [Coronel (11-3-6)]
|
[AY302550]
|
(E-26)
|
|
|
Echovirus 27 [Bacon (1-36-4)]
|
[AY302551]
|
(E-27)
|
|
|
Echovirus 29 [JV-10]
|
[AY302552]
|
(E-29)
|
|
|
Echovirus 30 [Bastianni]
|
[AF102711, AF311938]
|
(E-30)
|
|
|
Echovirus 31 [Caldwell]
|
[AY302554]
|
(E-31)
|
|
|
Echovirus 32 [PR-10]
|
[AY302555]
|
(E-32)
|
|
|
Echovirus 33 [Toluca-3]
|
[AY302556]
|
(E-33)
|
|
|
Enterovirus B69 [Toluca-1]
|
[AY302560]
|
(EV-B69)
|
|
|
Enterovirus B73 [USA/CA55-1988]
|
[AF241359]
|
(EV-B73)
|
|
|
Enterovirus B74 [USA/CA75-10213]
|
[AY556057]
|
(EV-B74)
|
|
|
Enterovirus B75 [OK85-10362]
|
[AY556070]
|
(EV-B75)
|
|
|
Enterovirus B77 [CF496-99]
|
[AJ493062]
|
(EV-B77)
|
|
|
Enterovirus B78 [Agius; W137-126/99]
|
[AY208120]
|
(EV-B78)
|
|
|
Enterovirus B79 [CA79-10384]
|
[AY843297]
|
(EV-B79)
|
|
|
Enterovirus B80 [CA67-10387]
|
[AY843298]
|
(EV-B80)
|
|
|
Enterovirus B81 [CA68-10389]
|
[AY843299]
|
(EV-B81)
|
|
|
Enterovirus B82 [CA64-10390]
|
[AY843300]
|
(EV-B82)
|
|
|
Enterovirus B83 [CA76-10392]
|
[AY843301]
|
(EV-B83)
|
|
|
Enterovirus B84 [CIV2003-10603]
|
[DQ902712]
|
(EV-B84)
|
|
|
Enterovirus B85 [BAN00-10353]
|
[AY843303]
|
(EV-B85)
|
|
|
Enterovirus B86 [BAN00-10354]
|
[AY843304]
|
(EV-B86)
|
|
|
Enterovirus B87 [BAN01-10396]
|
[AY843305]
|
(EV-B87)
|
|
|
Enterovirus B88 [BAN01-10398]
|
[AY843306]
|
(EV-B88)
|
|
|
Enterovirus B93 [38-03 (DR Congo)]
|
[EF127244]
|
(EV-B93)
|
|
|
Enterovirus B97 [BAN99-10355]
|
[AY843307]
|
(EV-B97)
|
|
|
Enterovirus B98 [T92-1499]
|
[AB426608]
|
(EV-B98)
|
|
|
Enterovirus B100 [BAN2000-10500]
|
[DQ902713]
|
(EV-B100)
|
|
|
Enterovirus B101 [CIV03-10361]
|
[AY843308]
|
(EV-B101)
|
|
|
Enterovirus B106 [BAN2001-10634]
|
|
(EV-B106)
|
|
|
Enterovirus B107 [TN94-0349]
|
[AB426609]
|
(EV-B107)
|
|
|
Enterovirus B110 [LM1861]
|
|
(EV-B110)
|
|
|
Simian agent 5 [B165]
|
[AF326751]
|
(SA5)
|
|
Human enterovirus C
|
|
|
|
|
|
Poliovirus 1 [Mahoney]
|
[J02281]
|
(PV-1)
|
|
|
Poliovirus 2 [Lansing]
|
[M12197]
|
(PV-2)
|
|
|
Poliovirus 3 [Leon]
|
[K01392]
|
(PV-3)
|
|
|
Coxsackievirus A1 [Tompkins]
|
[AF499035]
|
(CV-A1)
|
|
|
Coxsackievirus A11 [Belgium 1]
|
[AF499636]
|
(CV-A11)
|
|
|
Coxsackievirus A13 [Flores]
|
[AF499637, AF465511]
|
(CV-A13)
|
|
|
Coxsackievirus A17 [G-12]
|
[AF499639]
|
(CV-A17)
|
|
|
Coxsackievirus A19 [NIH-8663 (Dohi)]
|
[AF499641]
|
(CV-A19)
|
|
|
Coxsackievirus A20 [IH-35]
|
[AF499642, AF465514]
|
(CV-A20)
|
|
|
Coxsackievirus A21 [Kuykendall]
|
[AF546702, AF465515]
|
(CV-A21)
|
|
|
Coxsackievirus A22 [Chulman]
|
[AF499643]
|
(CV-A22)
|
|
|
Coxsackievirus A24 [Joseph]
|
[EF026081]
|
(CV-A24)
|
|
|
Enterovirus C95 [5-05]
|
|
(EV-C95)
|
|
|
Enterovirus C96 [BAN00-10488]
|
[EF015886]
|
(EV-C96)
|
|
|
Enterovirus C99 [USA/GA84-10636]
|
[EF555644]
|
(EV-C99)
|
|
|
Enterovirus C102 [BAN99-10424]
|
[EF555645]
|
(EV-C102)
|
|
|
Enterovirus C104 [CL-1231094 FL]
|
[EU840733]
|
(EV-C104)
|
|
|
Enterovirus C105 [TW/NTU07-1841]
|
|
(EV-C105)
|
|
|
Enterovirus C109 [NICA08-4327]
|
[GQ865517]
|
(EV-C109)
|
|
|
Enterovirus C113 [BAN-11609]
|
|
(EV-C113)
|
|
Human enterovirus D
|
|
|
|
|
|
Enterovirus D68 [Fermon]
|
[AY426531]
|
(EV-D68)
|
|
|
Enterovirus D70 [J670/71]
|
[D00820, DQ201177]
|
(EV-D70)
|
|
|
Enterovirus D94 [E210]
|
[DQ916376]
|
(EV-D94)
|
|
|
Enterovirus D111 [KK2640]
|
|
(EV-D111)
|
|
Porcine enterovirus B
|
|
|
|
|
|
Porcine enterovirus 9 [UKG/410/73]
|
[AF363453]
|
(PEV-9)
|
|
|
Porcine enterovirus 10 [LP54/England/75]
|
[AF363455]
|
(PEV-10)
|
|
Simian enterovirus A
|
|
|
|
|
|
Simian enterovirus A1 [SV4-1715 UWB]
|
[AF326759]
|
(SEV-A1)
|
|
|
(Simian virus 4, simian virus 28, simian agent 4)
|
|
|
|
Human rhinovirus A
|
|
|
|
|
|
Human rhinovirus A1 [2060]
|
[FJ445111]
|
(HRV-A1)
|
|
|
Human rhinovirus A2 [HGP]
|
[X02316]
|
(HRV-A2)
|
|
|
Human rhinovirus A7 [68-CV11]
|
[FJ445176; DQ473503]
|
(HRV-A7)
|
|
|
Human rhinovirus A8 [MRH-CV12]
|
[FJ445113]
|
(HRV-A8)
|
|
|
Human rhinovirus A9 [211-CV13]
|
[FJ445177]
|
(HRV-A9)
|
|
|
Human rhinovirus A10 [204-CV14]
|
[FJ445178; DQ473498]
|
(HRV-A10)
|
|
|
Human rhinovirus A11 [1-CV15]
|
[EF173414]
|
(HRV-A11)
|
|
|
Human rhinovirus A12 [181-CV16]
|
[EF173415]
|
(HRV-A12)
|
|
|
Human rhinovirus A13 [353]
|
[FJ445116]
|
(HRV-A13)
|
|
|
Human rhinovirus A15 [1734]
|
[DQ473493]
|
(HRV-A15)
|
|
|
Human rhinovirus A16 [11757]
|
[L24917]
|
(HRV-A16)
|
|
|
Human rhinovirus A18 [5986-CV17]
|
[FJ445118]
|
(HRV-A18)
|
|
|
Human rhinovirus A19 [6072-CV18]
|
[FJ445119]
|
(HRV-A19)
|
|
|
Human rhinovirus A20 [15-CV19]
|
[FJ445120]
|
(HRV-A20)
|
|
|
Human rhinovirus A21 [47-CV21]
|
[FJ445121]
|
(HRV-A21)
|
|
|
Human rhinovirus A22 [127-CV22]
|
[FJ445122]
|
(HRV-A22)
|
|
|
Human rhinovirus A23 [5124-CV24]
|
[DQ473497]
|
(HRV-A23)
|
|
|
Human rhinovirus A24 [5146-CV25]
|
[FJ445190; EF173416]
|
(HRV-A24)
|
|
|
Human rhinovirus A25 [5426-CV26]
|
[FJ445123]
|
(HRV-A25)
|
|
|
Human rhinovirus A28 [6101-CV29]
|
[DQ473508]
|
(HRV-A28)
|
|
|
Human rhinovirus A29 [5582-CV30]
|
[FJ445125]
|
(HRV-A29)
|
|
|
Human rhinovirus A30 [106F]
|
[FJ445179; DQ473512]
|
(HRV-A30)
|
|
|
Human rhinovirus A31 [140F]
|
[FJ445126]
|
(HRV-A31)
|
|
|
Human rhinovirus A32 [363]
|
[FJ445127]
|
(HRV-A32)
|
|
|
Human rhinovirus A33 [1200 ]
|
[FJ445128]
|
(HRV-A33)
|
|
|
Human rhinovirus A34 [137-3]
|
[FJ445189; DQ473501]
|
(HRV-A34)
|
|
|
Human rhinovirus A36 [342H]
|
[DQ473505]
|
(HRV-A36)
|
|
|
Human rhinovirus A38 [CH79]
|
[FJ445180; DQ473495]
|
(HRV-A38)
|
|
|
Human rhinovirus A39 [209]
|
[AY751783]
|
(HRV-A39)
|
|
|
Human rhinovirus A40 [1794]
|
[FJ445129]
|
(HRV-A40)
|
|
|
Human rhinovirus A41 [56110]
|
[DQ473491]
|
(HRV-A41)
|
|
|
Human rhinovirus A43 [58750]
|
[FJ445131]
|
(HRV-A43)
|
|
|
Human rhinovirus A44 [71560]
|
[DQ473499]
|
(HRV-A44)
|
|
|
Human rhinovirus A45 [Tippett (Baylor 1)]
|
[FJ445132]
|
(HRV-A45)
|
|
|
Human rhinovirus A46 [Crell (Baylor 2)]
|
[DQ473506]
|
(HRV-A46)
|
|
|
Human rhinovirus A47 [Calvert (Baylor 3)]
|
[FJ445133]
|
(HRV-A47)
|
|
|
Human rhinovirus A49 [8213]
|
[DQ473496]
|
(HRV-A49)
|
|
|
Human rhinovirus A50 [A2-58]
|
[FJ445135]
|
(HRV-A50)
|
|
|
Human rhinovirus A51 [F01-4081]
|
[FJ445136]
|
(HRV-A51)
|
|
|
Human rhinovirus A53 [F01-3928]
|
[DQ473507]
|
(HRV-A53)
|
|
|
Human rhinovirus A54 [F01-3774]
|
[FJ445138]
|
(HRV-A54)
|
|
|
Human rhinovirus A55 [Wis315E]
|
[DQ473511]
|
(HRV-A55)
|
|
|
Human rhinovirus A56 [CH82 [V-151-011-021]]
|
[FJ445140]
|
(HRV-A56)
|
|
|
Human rhinovirus A57 [CH47]
|
[FJ445141]
|
(HRV-A57)
|
|
|
Human rhinovirus A58 [21-CV20]
|
[FJ445142]
|
(HRV-A58)
|
|
|
Human rhinovirus A59 [611-CV35]
|
[DQ473500]
|
(HRV-A59)
|
|
|
Human rhinovirus A60 [2268-CV37]
|
[FJ445143]
|
(HRV-A60)
|
|
|
Human rhinovirus A61 [6669-CV39]
|
[FJ445144]
|
(HRV-A61)
|
|
|
Human rhinovirus A62 [1963M-CV40]
|
[FJ445145]
|
(HRV-A62)
|
|
|
Human rhinovirus A63 [6360-CV41]
|
[FJ445146]
|
(HRV-A63)
|
|
|
Human rhinovirus A64 [6258-CV44]
|
[FJ445181; EF173417]
|
(HRV-A64)
|
|
|
Human rhinovirus A65 [425-CV47]
|
[FJ445147]
|
(HRV-A65)
|
|
|
Human rhinovirus A66 [1983-CV48]
|
[FJ445148]
|
(HRV-A66)
|
|
|
Human rhinovirus A67 [1857-CV51]
|
[FJ445149]
|
(HRV-A67)
|
|
|
Human rhinovirus A68 [F02-2317 Wood]
|
[FJ445150]
|
(HRV-A68)
|
|
|
Human rhinovirus A71 [SF365]
|
[FJ445152]
|
(HRV-A71)
|
|
|
Human rhinovirus A73 [107E]
|
[DQ473492]
|
(HRV-A73)
|
|
|
Human rhinovirus A74 [328A]
|
[DQ473494]
|
(HRV-A74)
|
|
|
Human rhinovirus A75 [328F]
|
[DQ473510]
|
(HRV-A75)
|
|
|
Human rhinovirus A76 [H00062]
|
[FJ445182; DQ473502]
|
(HRV-A76)
|
|
|
Human rhinovirus A77 [130-63]
|
[FJ445154]
|
(HRV-A77)
|
|
|
Human rhinovirus A78 [2030-65]
|
[FJ445183; EF173418]
|
(HRV-A78)
|
|
|
Human rhinovirus A80 [277G]
|
[FJ445156]
|
(HRV-A80)
|
|
|
Human rhinovirus A81 [483F2]
|
[FJ445157]
|
(HRV-A81)
|
|
|
Human rhinovirus A82 [03647]
|
[FJ445160]
|
(HRV-A82)
|
|
|
Human rhinovirus A85 [50-525-CV54]
|
[FJ445163]
|
(HRV-A85)
|
|
|
Human rhinovirus A88 [CVD 01-0165-Dambrauskas]
|
[DQ473504]
|
(HRV-A88)
|
|
|
Human rhinovirus A89 [41467-Gallo]
|
[FJ445184; M16248]
|
(HRV-A89)
|
|
|
Human rhinovirus A90 [K2305]
|
[FJ445167]
|
(HRV-A90)
|
|
|
Human rhinovirus A94 [SF-1803]
|
[FJ445185; EF173419]
|
(HRV-A94)
|
|
|
Human rhinovirus A95 [SF-998]
|
[FJ445170]
|
(HRV-A95)
|
|
|
Human rhinovirus A96 [SF-1426]
|
[FJ445171]
|
(HRV-A96)
|
|
|
Human rhinovirus A98 [SF-4006]
|
[FJ445173]
|
(HRV-A98)
|
|
|
Human rhinovirus A100 [K6579]
|
[FJ445175]
|
(HRV-A100)
|
|
|
Human rhinovirus A101 [hrv-A101]
|
[GQ415051]
|
(HRV-A101)
|
|
Human rhinovirus B
|
|
|
|
|
|
Human rhinovirus B3 [FEB]
|
[DQ473485; EF173422]
|
(HRV-B3)
|
|
|
Human rhinovirus B4 [16/60]
|
[DQ473490]
|
(HRV-B4)
|
|
|
Human rhinovirus B5 [Norman]
|
[FJ445112]
|
(HRV-B5)
|
|
|
Human rhinovirus B6 [Thompson]
|
[DQ473486]
|
(HRV-B6)
|
|
|
Human rhinovirus B14 [1059]
|
[K02121, L05355]
|
(HRV-B14)
|
|
|
Human rhinovirus B17 [33342]
|
[EF173420]
|
(HRV-B17)
|
|
|
Human rhinovirus B26 [5660-CV27]
|
[FJ445124]
|
(HRV-B26)
|
|
|
Human rhinovirus B27 [5870-CV28]
|
[FJ445186; EF173421]
|
(HRV-B27)
|
|
|
Human rhinovirus B35 [164A]
|
[DQ473487]
|
(HRV-B35)
|
|
|
Human rhinovirus B37 [151-1]
|
[EF173423]
|
(HRV-B37)
|
|
|
Human rhinovirus B42 [56822]
|
[FJ445130]
|
(HRV-B42)
|
|
|
Human rhinovirus B48 [1505]
|
[DQ473488]
|
(HRV-B48)
|
|
|
Human rhinovirus B52 [F01-3772]
|
[FJ445188; EF173424]
|
(HRV-B52)
|
|
|
Human rhinovirus B69 [F02-2513-Mitchinson]
|
[FJ445151]
|
(HRV-B69)
|
|
|
Human rhinovirus B70 [F02-2547-Treganza]
|
[DQ473489]
|
(HRV-B70)
|
|
|
Human rhinovirus B72 [K2207]
|
[FJ445153]
|
(HRV-B72)
|
|
|
Human rhinovirus B79 [101-1]
|
[FJ445155]
|
(HRV-B79)
|
|
|
Human rhinovirus B83 [Baylor 7]
|
[FJ445161]
|
(HRV-B83)
|
|
|
Human rhinovirus B84 [432D]
|
[FJ445162]
|
(HRV-B84)
|
|
|
Human rhinovirus B86 [121564-Johnson]
|
[FJ445164]
|
(HRV-B86)
|
|
|
Human rhinovirus B91 [JM1]
|
[FJ445168]
|
(HRV-B91)
|
|
|
Human rhinovirus B92 [SF-1662]
|
[FJ445169]
|
(HRV-B92)
|
|
|
Human rhinovirus B93 [SF-1492]
|
[EF173425]
|
(HRV-B93)
|
|
|
Human rhinovirus B97 [SF-1372]
|
[FJ445172]
|
(HRV-B97)
|
|
|
Human rhinovirus B99 [604]
|
[FJ445174]
|
(HRV-B99)
|
|
Human rhinovirus C
|
|
|
|
|
|
Human rhinovirus C1 [NAT001 ]
|
[EF077279]
|
(HRV-C1 )
|
|
|
Human rhinovirus C2 [NAT045 ]
|
[EF077280]
|
(HRV-C2 )
|
|
|
Human rhinovirus C3 [QPM ]
|
[EF186077]
|
(HRV-C3 )
|
|
|
Human rhinovirus C4 [024]
|
[EF582385]
|
(HRV-C4 )
|
|
|
Human rhinovirus C5 [025]
|
[EF582386]
|
(HRV-C5 )
|
|
|
Human rhinovirus C6 [026]
|
[EF582387]
|
(HRV-C6 )
|
|
|
Human rhinovirus C7 [NY-074 ]
|
[DQ875932]
|
(HRV-C7 )
|
|
|
Human rhinovirus C8 [N4 ]
|
[GQ223227]
|
(HRV-C8 )
|
|
|
Human rhinovirus C9 [N10 ]
|
[GQ223228]
|
(HRV-C9 )
|
|
|
Human rhinovirus C10 [QCE ]
|
[GQ323774]
|
(HRV-C10 )
|
|
|
Human rhinovirus C11 [CL-170085 ]
|
[EU840952]
|
(HRV-C11 )
|
|
|
Human rhinovirus C12 [Resp_3922/07]
|
[HM236958]
|
(HRV-C12 )
|
|
|
Human rhinovirus C13 [Resp_2951/06]
|
[HM236908]
|
(HRV-C13 )
|
|
|
Human rhinovirus C14 [Resp_3090/06]
|
[HM236911]
|
(HRV-C14 )
|
|
|
Human rhinovirus C15 [Resp_4644/07]
|
[HM236963]
|
(HRV-C15 )
|
|
|
Human rhinovirus C16 [Resp_5910/07]
|
[HM236944]
|
(HRV-C16 )
|
|
|
Human rhinovirus C17 [Resp_5145/07]
|
[HM236936]
|
(HRV-C17 )
|
|
|
Human rhinovirus C18 [Resp_3631/07]
|
[HM236918]
|
(HRV-C18 )
|
|
|
Human rhinovirus C19 [CL-Fnp5]
|
[EU840728]
|
(HRV-C19 )
|
|
|
Human rhinovirus C20 [Resp_3995/07]
|
[HM236923]
|
(HRV-C20 )
|
|
|
Human rhinovirus C21 [Resp_5071/07]
|
[HM236903]
|
(HRV-C21 )
|
|
|
Human rhinovirus C22 [Resp_2748/06]
|
[HM236905]
|
(HRV-C22 )
|
|
|
Human rhinovirus C23 [Resp_3053/06]
|
[HM236901]
|
(HRV-C23 )
|
|
|
Human rhinovirus C24 [Resp_7147/07]
|
[HM236939]
|
(HRV-C24 )
|
|
|
Human rhinovirus C25 [Resp_2832/06]
|
[HM236952]
|
(HRV-C25 )
|
|
|
Human rhinovirus C26 [Resp_2514/06]
|
[HM236904]
|
(HRV-C26 )
|
|
|
Human rhinovirus C27 [Resp_2784/06]
|
[HM236906]
|
(HRV-C27 )
|
|
|
Human rhinovirus C28 [Resp_3105/06]
|
[HM236954]
|
(HRV-C28 )
|
|
|
Human rhinovirus C29 [Resp_5345/07]
|
[HM236949]
|
(HRV-C29 )
|
|
|
Human rhinovirus C30 [Resp_3898/07]
|
[HM236968]
|
(HRV-C30 )
|
|
|
Human rhinovirus C31 [Resp_4923/07]
|
[HM236964]
|
(HRV-C31 )
|
|
|
Human rhinovirus C32 [Resp_6131/07]
|
[HM236897]
|
(HRV-C32 )
|
|
|
Human rhinovirus C33 [Resp_4917/07]
|
[HM236934]
|
(HRV-C33 )
|
|
|
Human rhinovirus C34 [pico58]
|
|
(HRV-C34)
|
|
|
Human rhinovirus C35 [lrti_hrvc1]
|
|
(HRV-C35)
|
|
|
Human rhinovirus C36 [Resp_2480/07]
|
[JF416311]
|
(HRV-C36)
|
|
|
Human rhinovirus C37 [Resp_6135/08]
|
[JF416321; GU294476]
|
(HRV-C37)
|
|
|
Human rhinovirus C38 [Resp_6142/08]
|
[JF416322; GU294477]
|
(HRV-C38)
|
|
|
Human rhinovirus C39 [Resp_10221/08]
|
[JF416306]
|
(HRV-C39)
|
|
|
Human rhinovirus C40 [Resp_2800/06]
|
[JF416312]
|
(HRV-C40)
|
|
|
Human rhinovirus C41 [Resp_9449/08]
|
[JF416323]
|
(HRV-C41)
|
|
|
Human rhinovirus C42 [Resp_5477/08]
|
[JF416320]
|
(HRV-C42)
|
|
|
Human rhinovirus C43 [Resp_13229/08]
|
[JF416307]
|
(HRV-C43)
|
|
|
Human rhinovirus C44 [Resp_15588/09]
|
[JF416310]
|
(HRV-C44)
|
|
|
Human rhinovirus C45 [Resp_13958/08]
|
[JF416308]
|
(HRV-C45)
|
|
|
Human rhinovirus C46 [Resp_5153/07]
|
[JF416318; GU294446]
|
(HRV-C46)
|
|
|
Human rhinovirus C47 [PNG7254-3947]
|
[JF519760; JF519761 ]
|
(HRV-C47)
|
|
|
Human rhinovirus C48 [PNG7293-3193]
|
[JF519762; JF519763]
|
(HRV-C48)
|
Species names are in italic script. Beneath each species name is listed a representative isolate in roman script, with sequence accession number [ ]. The names of any additional strains are also listed, abbreviated as follows: bovine enterovirus (BEV); coxsackievirus (CV); echovirus (E); enterovirus (EV); human rhinovirus (HRV); poliovirus (PV); porcine enterovirus (PEV); simian agent (SA); simian virus (SV); simian enterovirus (SEV).
List of other related viruses which may be members of the genus Enterovirus but have not been approved as species
|
Enterovirus 103 [USA/GA99-POo-1] |
[FJ007373] |
(EV-103) |
|
Enterovirus 108 [N125] |
[AF414372] |
(EV-108) |
|
Enterovirus 112 [BAN-11217] |
|
(EV-112) |
|
Enterovirus 115 [BAN-11617] |
|
(EV-115) |
|
Simian virus 6 [1631] |
[AF326766] |
(SV6) |
|
Simian virus 47 [OM107] |
|
(SV47) |
Genus Cardiovirus
Type species Encephalomyocarditis virus
Virion properties
Morphology
Empty capsids are seen only rarely, if ever. When compared by mean wall thickness, surface unevenness and chain length of the major proteins, the cardiovirus capsid is intermediate between the enteroviruses and aphthoviruses. In place of a continuous, circular, canyon, seen in enteroviruses, is a five-fold repeated pit. There is no pocket factor.
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.33–1.34 g cm−3. Virions are moderately stable to acidic pH.
Nucleic acid
EMCV has a poly(C) tract of variable length (usually 80–250 nt) about 150 nt from the 5′-terminus of the viral RNA, while theilovirus isolates lack this feature. All EMCV members have two pseudoknots 5′ to their poly(C) tracts. The IRES is of type II. The cre is located in the 1B region of both cardiovirus species. The nt sequence identity over the entire genome for different species of the genus Cardiovirus is more than 50% (e.g. TMEV has 54% nt sequence identity to EMCV).
Genome organization and replication
Genome layout:
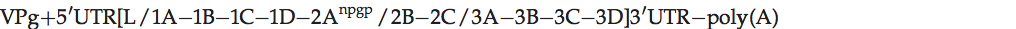
The viral genome encodes a leader (L) protein which lacks proteolytic activity, unlike the L of aphthoviruses; thus L is cleaved from P1 by the virus encoded protease 3C. The 1D/2A junction is also cleaved by 3Cpro, rather than by 2A. The 2A protein causes polypeptide chain interruption, between P1-2A and downstream sequences at an essential sequence, NPG↓P.
Antigenic properties
Four independent antigenic sites have been described. There is no evidence of an N-D conversion, nor of “A” particles. The species Encephalomyocarditis virus consists of a single serotype. However, the species Theilovirus consists of 12 genetic types, Theiler’s murine encephalomyelitis virus (TMEV), Vilyuisk human encephalomyelitis virus (VHEV), thera virus (formerly named Theiler-like virus of rats), and Saffold virus (SAFV) types 1 to 9. There is no cross-neutralization between TMEV and VHEV; however, the antigenic relationships between these and thera and saffold viruses is presently not known.
Biological properties
Encephalomyocarditis viruses have been isolated from over 30 host species, including mammals, birds and invertebrates. Clinical manifestations include encephalitis and myocarditis in mice and many other animals. TMEV can be divided into two biological subgroups which both infect mice; one causes an acute and fatal polioencephalomyelitis and the other causes a chronic persistent demyelinating infection of the white matter. VHEV was isolated (in mice) from a person suffering from a degenerative neurological disease (Vilyuisk encephalitis), however, since the virus was extensively passaged in mice during the 1950s it is not clear if it is of human or mouse origin. Thera virus has been isolated from clinically normal rats. Saffold viruses have been isolated from humans, especially children, and have been associated with both respiratory disease and gastroenteritis. Cardiovirus infection does not cause cleavage of the host eIF-4G. The cellular receptor used by EMCV to attach to murine vascular endothelial cells has been identified as VCAM-1. However, in human cell lines an as yet unidentified sialoglycoprotein(s) has been found. EMCV binds to human erythrocytes via glycophorin A. Low-neurovirulence TMEVs use sialic acid to attach to mammalian cells, while glycosaminoglycan heparan sulphate is involved in the attachment of high-neurovirulence TMEVs.
Species demarcation criteria in the genus
Members of a species of the genus Cardiovirus:
- share greater than 70% aa identity in the polyprotein
- share greater than 60% aa identity in P1
- share greater than 70% aa identity in 2C + 3CD
- share a natural host range
- share a common genome organization.
List of species in the genus Cardiovirus
|
Encephalomyocarditis virus |
|
|
|
|
|
Encephalomyocarditis virus 1 [R] |
[M81861] |
(EMCV-1) |
|
|
(Mengovirus, Columbia SK virus and Maus Elberfeld virus are all strains of EMCV-1) |
|
|
|
Theilovirus |
|
|
|
|
|
Theiler’s murine encephalomyelitis virus [GD VII] |
[M20562, X56019] |
(TMEV) |
|
|
Vilyuisk human encephalomyelitis virus [V1/Siberia/55] |
[M80888, M94868, EU723237] |
(VHEV) |
|
|
Thera virus [NGS910] |
[AB090161] |
(TRV) |
|
|
(Theiler-like virus of rats) |
|
|
|
|
Saffold virus 1 [California/81] |
[EF165067] |
(SAFV-1) |
|
|
Saffold virus 2 [Can112051-06] |
[AM922293] |
(SAFV-2) |
|
|
Saffold virus 3 [D/VI2273/2004] |
[EU681178] |
(SAFV-3) |
|
|
Saffold virus 4 [Pak5842] |
[FJ463606; FJ463611] |
(SAFV-4) |
|
|
Saffold virus 5 [Pak5003] |
[FJ463615] |
(SAFV-5) |
|
|
Saffold virus 6 [Pak6572] |
[FJ463617] |
(SAFV-6) |
|
|
Saffold virus 7 [Afg1449] |
[FJ463602, FJ463613] |
(SAFV-7) |
|
|
Saffold virus 8 [Pak1141] |
[FJ463604, FJ463614] |
(SAFV-8) |
|
|
Saffold virus 9 [Nig239] |
[FJ997541, FJ997532, FJ997535] |
(SAFV-9) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cardiovirus but have not been approved as species
None reported.
Genus Aphthovirus
Type species Foot-and-mouth disease virus
Virion properties
Morphology
The capsid of FMDV is thin-walled (mean thickness ca. 33 Å) and has an unusually smooth surface. A long (17–23 aa), mobile loop, the G-H loop, projects from the surface of 1D. There is a pore at the five-fold axis, where part of the underlying 1C is exposed. Some serotypes of FMDV accumulate empty capsids.
Physicochemical and physical properties
Virions are acid-labile; FMDV particles are unstable below pH 6.8; equine rhinitis A virus (ERAV) particles are unstable below pH 5.5. The buoyant density in CsCl is 1.43–1.45 g cm−3. Virions of FMDV sediment at 146S, empty capsids at 75S.
Nucleic acid
There is a poly(C) tract close to the 5’ terminus of the genome. In FMDV it is located about 360 nt from the end, and varies in length from 100 to more than 400 nt. Current data suggest that the poly(C) tract in ERAV is shorter (ca. 40 nt) and closer to the 5’ end. In the RNA of FMDV there is a series of 3–4 pseudoknots on the 3’-side of the poly(C); in ERAV these pseudoknots are formed by perfectly repeated sequences each consisting of 21 bases; the total 5’-UTR is thus extremely long (1.1–1.5 kb). No pseudoknots have so far been identified in the bovine rhinitis viruses. The IRES is of type II. The FMDV cre is located in the 5’ UTR between the repeated pseudoknots and the IRES, but has not been identified for the other aphthovirus species. ERAV and FMDV differ by approximately 50% in nt sequence across the entire genome.
Genome organization and replication
Genome layout:
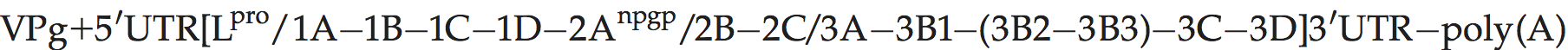
Translation starts at two alternative in-frame initiation sites, resulting in two forms of the L protein (Lab and Lb). L is a papain-like cysteine protease which cleaves itself from the virus polyprotein. The 2A polypeptide is very short (chain length = 18 aa in FMDV), and is involved in NPGP-dependent polypeptide chain interruption at its C-terminus as in cardioviruses. The genome of FMDV encodes three species of VPg while those of ERAV, BRAV and BRBV encode only one.
Antigenic properties
Five independent antigenic sites have been reported in FMDV type O, two of which have determinants in the G-H loop of 1D. There is no evidence of N-D conversion, nor “A” particles.
Biological properties
This genus is comprised of viruses which primarily infect via the upper respiratory tract. FMDV infects mainly cloven-hoofed animals, but has been isolated from at least 70 species of mammals. Clinical manifestations of FMDV infections include foot-and-mouth disease (vesicular lesions), sometimes with associated acute fatal myocarditis in young animals. ERAV causes upper respiratory tract infections of horses, but may infect a number of other species including man. Bovine rhinitis A viruses (BRAV) and bovine rhinitis B virus (BRBV) infect the respiratory tract of cattle. FMDV and ERAV may produce persistent upper respiratory tract infections. FMDV infects cells by binding to integral membrane proteins of the integrin family through its 1D G-H loop; the principle integrin used is αvβ6. Heparan sulphate proteoglycans may also serve as receptors in cell cultures and at least one other unidentified receptor has been proposed. ERAV can use sialic acid to bind to cells. Cap-dependent translation of host mRNA is inhibited by Lpro, which cleaves the host eIF-4G.
Species demarcation criteria in the genus
Members of a species of the genus Aphthovirus:
- share greater than 70% aa identity in the polyprotein
- share greater than 60% aa identity in P1
- share greater than 80% aa identity in 2C + 3CD
- share a natural host range
- have a genome base composition which varies by no more than 1%
- share a common genome organization.
List of species in the genus Aphthovirus
|
Bovine rhinitis B virus |
|
|
|
|
|
Bovine rhinitis B virus 1 [EC 11] |
[EU236594] |
(BRBV-1) |
|
|
(Bovine rhinovirus 2) |
|
|
|
Equine rhinitis A virus |
|
|
|
|
|
Equine rhinitis A virus 1 [PERV] |
[DQ272578, X96870] |
(ERAV-1) |
|
|
(Equine rhinovirus 1) |
|
|
|
Foot-and-mouth disease virus |
|
|
|
|
|
Foot-and-mouth disease virus O [UK/1/24 (OV1)] |
[AY593829] |
(FMDV-O) |
|
|
Foot-and-mouth disease virus A [UK/119/32] |
[AY593752] |
(FMDV-A) |
|
|
Foot-and-mouth disease virus C [UK/149/34] |
[AY593810] |
(FMDV-C) |
|
|
Foot-and-mouth disease virus SAT 1 [RV/11/37] |
[AY593839] |
(FMDV-SAT1) |
|
|
Foot-and-mouth disease virus SAT 2 [RHO/1/48] |
[AY593847] |
(FMDV-SAT2) |
|
|
Foot-and-mouth disease virus SAT 3 [BEC/1/65] |
[AY593853] |
(FMDV-SAT3) |
|
|
Foot-and-mouth disease virus Asia 1 [PAK/1/54] |
[AY593795] |
(FMDV-Asia1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Aphthovirus but have not been approved as species
|
Bovine rhinitis A virus 1 [RS 3x] |
(BRAV-1) |
|
(Bovine rhinovirus 1) |
|
|
Bovine rhinitis A virus 2 [H-1] |
(BRAV-2) |
|
(Bovine rhinovirus 3) |
|
Genus Hepatovirus
Type species Hepatitis A virus
Distinguishing features
In contrast to those of other picornaviruses, protein 1A of hepatoviruses is extremely small, does not appear to be myristoylated at its N-terminus, and may not be a component of the mature virus particle. Immature HAV may contain uncleaved 1D2A (PX) precursor protein.
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
Viruses are very stable, resistant to acid pH and elevated temperatures (60 °C for 10 min). Buoyant density in CsCl is 1.32–1.34 g cm−3.
Nucleic acid
There is little similarity between the genome sequences of hepatoviruses and those of other picornaviruses. Although the IRES is distantly related to the type II IRES, it has been designated as type III. The 5′-UTR contains a 5′-terminal hairpin, two putative pseudoknots, and a short (ca. 40 nt) pyrimidine-rich (i.e. not a pure poly-C) tract upstream of the IRES. The cre is located in the 3D region. Nucleotide sequence identity between different hepatitis A virus (HAV) strains is generally greater than 80%. The G+C content of HAV genomes is unusually low at about 38%.
Genome organization and replication
Genome layout:
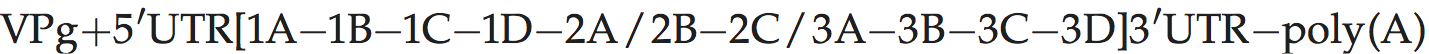
The polyprotein contains only a single protease (3Cpro). There is no clearly defined L protein, and 2A has no proteolytic activity. The primary cleavage of the polyprotein occurs at the 2A/2B junction, and is catalyzed by 3Cpro. The 1D/2A cleavage may be directed by an unknown cellular protease, or the VP1 protein may be subject to C-terminal trimming as in cardioviruses. Replication in cell culture occurs slowly, with little CPE, and with low yields of virus compared to most other picornaviruses. The IRES differs from those of other picornaviruses in that its activity is dependent on intact eIF-4G.
Antigenic properties
Hepatitis A viruses belong to a single serotype and are highly conserved in their antigenic properties. Most antibodies are directed against a single, conformationally defined immunodominant antigenic site that is comprised of aa residues of the VP3 and VP1 proteins on the surface of the virion.
Biological properties
HAV infects epithelial cells of the small intestine and hepatocytes of primates. Virus is predominantly replicated within the liver, excreted via the bile and present in feces in high titer. Viral shedding is maximal shortly before the onset of clinical signs of hepatitis, which probably represents immunopathologically-mediated liver injury. Clinical manifestations are fever, jaundice, light stools, abdominal pain, and occasionally diarrhea. HAV generally establishes a persistent infection when inoculated on to any of a wide range of primate cells in vitro, but persistent infection does not occur in vivo, and the viruses are not associated with chronic hepatitis. HAVs can be divided into two distinct biotypes that are phylogenetically distinct and have different preferred hosts (all species of primates: humans, chimpanzees, owl monkeys and marmosets, for one biotype, versus green monkeys and cynomolgus monkeys for the other). These two biotypes share cross-reacting antigens, but have biotype-specific epitopes that can be distinguished by monoclonal antibodies.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Hepatovirus
|
Hepatitis A virus |
|
|
|
Hepatitis A virus 1 [HM-175] |
[M14707] |
(HAV-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Hepatovirus but have not been approved as species
None reported.
Genus Parechovirus
Type species Human parechovirus
Distinguishing features
Predicted protein sequences of parechoviruses are highly divergent from other picornaviruses, no protein having a greater than 30% level of identity when compared with corresponding proteins of other genera. In contrast to most other picornaviruses, protein 1AB of parechoviruses appears not to be cleaved, and its N-terminus, also unusually, is not myristoylated. The mature capsid therefore appears to comprise only three proteins, 1AB, 1C and 1D.
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
Virions are acid-stable. The buoyant density in CsCl is 1.36 g cm−3.
Nucleic acid
The 5′-UTR is 710–730 nt and contains a typical type II IRES. The cre has been identified in the 1AB region for human parechoviruses and is thought to lie within the 3B region of Ljungan viruses. The ORF is 2180/2250 codons and the 3′-UTR 87 and 111 nt in human parechovirus and Ljungan virus, respectively.
Genome organization and replication
Genome layout:
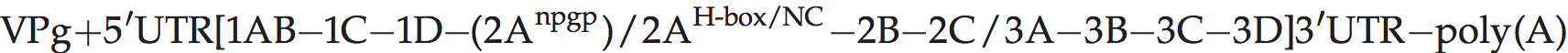
The polyprotein contains only a single protease (3Cpro). The 2A protein is believed to lack protease activity and is related distantly to a family of cellular proteins involved in the control of cell proliferation, as well as to that of kobuviruses and tremoviruses. Ljungan virus possesses an NPGP motif following the predicted 1D polypeptide, suggesting the possible presence of a second 2A; however, it is believed that this short 2A-like sequence may form part of 1D.
Antigenic properties
Human parechoviruses are divided into 14 genetic types and there is no cross-neutralization between types 1, 2 and 3. There may be a cross-reaction between types 2 and 5, however, the remaining 10 types have not been tested. Ljungan viruses are divided into four genetic types, but antigenic relationships have not been studied.
Biological properties
Human parechoviruses replicate in the respiratory and gastrointestinal tract. Infection is particularly prevalent in young children but it is probably often asymptomatic. In addition to respiratory infections and diarrhea, infections of the central nervous system have occasionally been reported. HPeV types 1 and 6 have been found in monkeys with diarrhea, although disease association was not proven. The cytopathology may be unusual in including changes in granularity and chromatin distribution in the nucleus, when viewed in the electron microscope. Ljungan viruses appear to infect predominantly rodents (voles) and have been proposed to infect humans; however, conclusive data are awaited. Some human parechoviruses (types 1, 2, 4, 5 and 6) possess an RGD tri-peptide (towards the carboxy-terminus of 1D) which is involved in integrin receptor-binding. The integrins αvβ3 and αvβ6 have been shown to be the primary receptors for HPeV-1 in A549 cells. Receptor usage for the remaining human parechoviruses and Ljungan viruses is not known.
Species demarcation criteria in the genus
Members of a species of the genus Parechovirus:
- share greater than 70% aa identity in the polyprotein
- share greater than 70% aa identity in P1
- share greater than 80% aa identity in 2C + 3CD
- share a natural host range
- share a common genome organization
- have a similar genome base composition which varies by no more than 1%.
List of species in the genus Parechovirus
|
Human Parechovirus |
|
|
|
|
|
Human parechovirus 1 [Harris] |
[L02971] |
(HPeV-1) |
|
|
(Human echovirus 22) |
|
|
|
|
Human parechovirus 2 [Williamson] |
[AJ005695] |
(HPeV-2) |
|
|
(Human echovirus 23) |
|
|
|
|
Human parechovirus 3 [A308/99] |
[AB084913] |
(HPeV-3) |
|
|
Human parechovirus 4 [K251176-02] |
[DQ315670] |
(HPeV-4) |
|
|
Human parechovirus 5 [CT86-6760] |
[AF055846] |
(HPeV-5) |
|
|
Human parechovirus 6 [NII561-2000] |
[AB252582] |
(HPeV-6) |
|
|
Human parechovirus 7 [PAK5045] |
[EU556224] |
(HPeV-7) |
|
|
Human parechovirus 8 [BR/217/2006] |
[EU716175] |
(HPeV-8) |
|
|
Human parechovirus 9 [BAN2004-10902] |
|
(HPeV-9) |
|
|
Human parechovirus 10 [BAN2004-10903] |
|
(HPeV-10) |
|
|
Human parechovirus 11 [BAN2004-10905] |
|
(HPeV-11) |
|
|
Human parechovirus 12 [BAN2004-10904] |
|
(HPeV-12) |
|
|
Human parechovirus 13 [BAN2005-10901] |
|
(HPeV-13) |
|
|
Human parechovirus 14 [451564 ] |
[FJ373179] |
(HPeV-14) |
|
|
Human parechovirus 15 [BAN-11614] |
|
(HPeV-15) |
|
|
Human parechovirus 16 [BAN-11615] |
|
(HPeV-16) |
|
Ljungan virus |
|
|
|
|
|
Ljungan virus 1 [87-012] |
[AF327920] |
(LV-1) |
|
|
Ljungan virus 2 [145SL] |
[AF327922] |
(LV-2) |
|
|
Ljungan virus 3 [M1146] |
[AF538689] |
(LV-3) |
|
|
Ljungan virus 4 [64-7855] |
[EU854568] |
(LV-4) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Parechovirus but have not been approved as species
None reported.
Genus Erbovirus
Type species Equine rhinitis B virus
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
Equine rhinitis B virus (ERBV) has a buoyant density in CsCl of 1.41–1.45 g cm−3. pH stability is variable; ERBV-1 and ERBV-2 are labile below pH 5.0 while ERBV-3 is stable over a wide pH range (2.2–8.0). ERBV is rapidly inactivated by heating at 50 °C but divalent cations stabilize against thermal inactivation.
Nucleic acid
ERBV possesses possibly the longest picornavirus genome, approaching 9 kb (the 5′ end has not been sequenced). The IRES is of type II, and a poly(C) tract is thought to be present. No pseudoknots have yet been identified within the 5′ UTR. The location of the cre has not been identified.
Proteins
The CPs have between 25% and 47% aa sequence identity to those of ERAV, FMDV and EMCV, although protein modelling studies indicate that the capsid of ERBV more closely resembles that of EMCV.
Genome organization and replication
Genome layout:
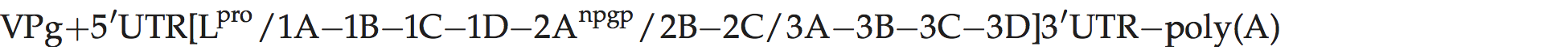
No evidence for alternative sites of initiation of protein synthesis is available, as is found in the aphthoviruses. The L protein appears to be a protease, but has only 23% and 18% aa sequence identity to the L proteins of FMDV and ERAV, respectively. The 2B and 3C proteins have exceptionally large chain lengths (283 and 251 aa). The 2A protein has a chain length of 18 aa, ending in NPG↓P, and there is only one VPg. The 3′-UTR is relatively long at 167 nt.
Antigenic properties
ERBV consists of three serotypes, ERBV-1, -2 and -3.
Biological properties
ERBV causes upper respiratory tract disease in horses, with a viremia and fecal shedding. Infections may be persistent.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Erbovirus
|
Equine rhinitis B virus |
|
|
|
|
|
Equine rhinitis B virus 1 [P1436/71] |
[X96871] |
(ERBV-1) |
|
|
(Equine rhinovirus 2) |
|
|
|
|
Equine rhinitis B virus 2 [P313/75] |
[AF361253] |
(ERBV-2) |
|
|
(Equine rhinovirus 3) |
|
|
|
|
Equine rhinitis B virus 3 [4442/75 ] |
[DQ108383] |
(ERBV-3) |
|
|
(Acid-stable equine picornavirus) |
|
|
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Erbovirus but have not been approved as species
None reported.
Genus Kobuvirus
Type species Aichi virus
Distinguishing features
Protein 1AB appears not to be cleaved; however, a myristoylation signal (GxxxT) is present at the amino terminus of the polypeptide.
Virion properties
Morphology
Unlike other picornaviruses, kobuvirus capsids show a distinctive surface morphology when observed by electron microscopy (Figure 1h).
Physicochemical and physical properties
Virions are stable at pH 3.5.
Nucleic acid
The genome of Aichi virus (AiV) has a high G+C base composition (59%), and a very long 3′-UTR (240 nt), however, the 3′-UTRs of bovine kobuvirus (BKV) and porcine kobuvirus (PKV) are shorter at 177 and 170 nt, respectively. In AiV-1, there is a 5′-proximal stem-loop involved in RNA replication and encapsidation. The IRES of AiV-1 and BKV-1 is of type II, however, PKV (which has not yet been assigned to a species) possesses a type IV IRES. The location of the cre has not been identified.
Genome organization and replication
Genome layout:
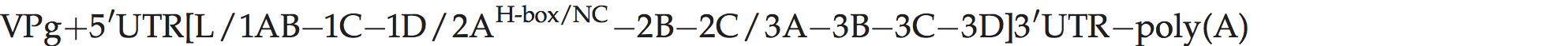
There is a leader polypeptide of unknown function, and distinctive length (170–195 aa rather than 67 aa or 217 aa in EMCV and FMDV, respectively). The 2A protein contains an H-Box/NC motif and is distantly related to that of parechoviruses and tremoviruses.
Antigenic properties
The two kobuvirus species, Aichi virus and Bovine kobuvirus, each consists of a single serotype.
Biological properties
AiV grows in cell cultures (BSC-1, Vero). AiV is thought to be a cause of human gastroenteritis. BKV has been isolated from cattle and sheep. PKV has been isolated from pigs.
Species demarcation criteria in the genus
Members of a species of the genus Kobuvirus:
- share greater than 70% aa identity in the polyprotein
- share greater than 70% aa identity in P1
- share greater than 80% aa identity in 2C + 3CD
- share a common genome organization.
List of species in the genus Kobuvirus
|
Aichi virus |
|
|
|
Aichi virus 1 [A846/88] |
[AB040749] |
(AiV-1) |
|
Bovine kobuvirus |
|
|
|
Bovine kobuvirus 1 [U-1] |
[AB084788] |
(BKV-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Kobuvirus but have not been approved as species
|
Porcine kobuvirus 1 [swine/S-1-HUN/2007/Hungary] |
[EU787450] |
(PKV-1) |
Genus Teschovirus
Type species Porcine teschovirus
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
Virions are stable at acid pH. Buoyant density in CsCl is 1.33 g cm−3. Empty capsids are often observed in virus preparations.
Nucleic acid
Teschoviruses have a type IV IRES about 290 nt in length which is functional in the absence of eIF-4G. In both these properties the IRES resembles that of hepatitis C virus (family Flaviviridae); sequence similarity has also been observed. The location of the cre is thought to be within the 2C region.
Proteins
Genomes encode a single VPg and a leader (L) protein. The 2A polypeptide is very short and ends in NPG↓P, indicative of an aphthovirus 2A-like molecule.
Genome organization and replication
Genome layout:
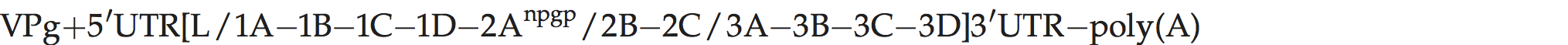
The genome layout is similar to that of the aphthoviruses, except that only a single VPg is present. The function of the leader polypeptide is unknown and is not predicted to have proteolytic activity.
Antigenic properties
Porcine teschoviruses are divided into 11 serotypes (PTV-1 to -11) which are distinct in cross-neutralization tests and can be differentiated using their 1D (VP1) sequences.
Biological properties
Clinical manifestations may include a polioencephalomyelitis (“Teschen/Talfan disease”, also known as teschovirus encephalomyelitis), which may vary in severity. The viruses have been associated with a number of disease syndromes, including reproductive and gastrointestinal disorders. The pig is the only known host.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Teschovirus
|
Porcine teschovirus |
|
|
|
|
|
Porcine teschovirus 1 [Talfan] |
[AF231769, AB038528] |
(PTV-1) |
|
|
(Porcine enterovirus 1) |
|
|
|
|
Porcine teschovirus 2 [T80] |
[AF296087] |
(PTV-2) |
|
|
(Porcine enterovirus 2) |
|
|
|
|
Porcine teschovirus 3 [O2b] |
[AF296088] |
(PTV-3) |
|
|
(Porcine enterovirus 3) |
|
|
|
|
Porcine teschovirus 4 [PS36] |
[AF296089] |
( PTV-4) |
|
|
(Porcine enterovirus 4) |
|
|
|
|
Porcine teschovirus 5 [F26] |
[AF296090] |
(PTV-5) |
|
|
(Porcine enterovirus 5) |
|
|
|
|
Porcine teschovirus 6 [PS37] |
[AF296091] |
(PTV-6) |
|
|
(Porcine enterovirus 6) |
|
|
|
|
Porcine teschovirus 7 [F43] |
[AF296092] |
(PTV-7) |
|
|
(Porcine enterovirus 7) |
|
|
|
|
Porcine teschovirus 8 [UKG/173/74] |
[AF296093] |
(PTV-8) |
|
|
(Porcine enterovirus 11) |
|
|
|
|
Porcine teschovirus 9 [Vir-2899/84] |
[AF296094] |
(PTV-9) |
|
|
(Porcine enterovirus 12) |
|
|
|
|
Porcine teschovirus 10 [Vir 461/88] |
[AF296119] |
(PTV-10) |
|
|
(Porcine enterovirus 13) |
|
|
|
|
Porcine teschovirus 11 [Dresden] |
[AF296096] |
(PTV-11) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Teschovirus but have not been approved as species
None reported.
Genus Sapelovirus
Type species Porcine sapelovirus
Distinguishing features
Sapeloviruses are most closely related to members of the Enterovirus genus, but possess a leader polypeptide of unknown function. The 2A polypeptide may be a cysteine protease, but it is distinct from that of the enteroviruses. The 2B and 3A proteins are also very different from the enteroviruses. In all three sapelovirus species the IRES is type IV, whereas in all enteroviruses species it is type I.
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
Viruses are very stable, resistant to acid pH and elevated temperatures (60 °C for 10 min). Buoyant density in CsCl is 1.32–1.34 g cm−3.
Nucleic acid
The IRES of all three sapelovirus species is type IV. The location of the cre has not been identified.
Genome organization and replication
Genome layout:
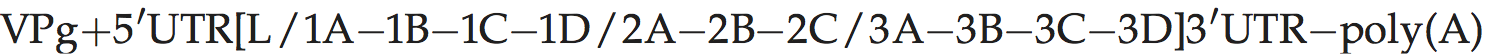
The predicted leader polypeptide of avian sapelovirus (ASV) is the longest known at 451 aa and suspected to be a trypsin-like protease. The leader polypeptide porcine sapelovirus (PSV) and simian sapelovirus (SSV) are much shorter at 84 aa and 88 aa, respectively. The 2A region of ASV is predicted to be mostly deleted with a residual 12 aa remaining. The 2A polypeptides of PSV and SSV are longer at 226 aa and 302 aa, respectively; these are both suspected to be a protease. The L/VP0 cleavage and presence of a potential myristoylation site on VP0 is predicted for ASV (kQ/GqvqS). However, the precise L/VP0 cleavage for PSV or SSV is not clear. The sequences in this region are quite well conserved between the three viruses but it would require unusual cleavages in PSV (qL/GqvhS) and SSV (qC/GqvqS) to generate a myristoylation signal. VP0 is cleaved to VP4 and VP2 as shown by N-terminal sequencing of the VP2 polypeptide for ASV and SSV.
Antigenic properties
Porcine sapelovirus consists of a single antigenically diverse serotype, PSV-1. Avian sapelovirus also consists of a single serotype, ASV-1. Simian sapelovirus consists of three genetically-defined types, SSV-1 to SSV-3.
Biological properties
Pigs, monkeys and ducks are the only known hosts for porcine, simian and avian sapeloviruses, respectively.
Species demarcation criteria in the genus
Members of a species of the genus Sapelovirus have:
- share greater than 70% aa identity in the polyprotein
- share greater than 64% aa identity in P1
- share greater than 70% aa identity in 2C+3CD
- a defined tissue tropism and host range
- a similar genome base composition which varies by no more than 1%
- a common genome organization.
List of species in the genus Sapelovirus
|
Porcine sapelovirus |
|
|
|
|
|
Porcine sapelovirus 1 [V13] |
[AF406813] |
(PSV-1) |
|
|
(Porcine enterovirus 8) |
|
|
|
Simian sapelovirus |
|
|
|
|
|
Simian sapelovirus 1 [SV2-2383] |
[AY064708] |
(SSV-1) |
|
|
(Simian virus 2) |
|
|
|
|
Simian sapelovirus 2 [SV49-2600] |
[AY064720, AF326765, AY064714] |
(SSV-2) |
|
|
(Simian virus 49) |
|
|
|
|
Simian sapelovirus 3 [SV16-2450-SD] |
[AY064715, AF326752, AY064709] |
(SSV-3) |
|
|
(Simian viruses 16, 18, 42, 44 & 45) |
|
|
|
Avian sapelovirus |
|
|
|
|
|
Avian sapelovirus 1 [TW90A] |
[AY563023] |
(ASV-1) |
|
|
(Duck picornavirus TW90A) |
|
|
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Sapelovirus but have not been approved as species
None reported.
Genus Senecavirus
Type species Seneca Valley virus
Virion properties
Morphology
No surface morphology is visible by EM. The X-ray crystallographic structure of Seneca Valley virus (SVV) has been determined at 2.3 Å resolution. The overall folds of the CPs are very similar to the corresponding proteins in other picornaviruses. Similar to cardioviruses, VP1 of SVV possesses a hydrophobic pocket without a pocket factor. However, the entrance to the hydrophobic cleft in SVV is almost completely sealed off by a number of VP1 residues. In cardioviruses the entrance is narrow while in the human rhinoviruses it is wide.
Physicochemical and physical properties
SVV is stable at pH 3.0.
Nucleic acid
The 5′ UTR is 666 nt long and contains a type IV IRES. The 3′ UTR is 71 nt long and the predicted folding of this region reveals two stem-loops with the potential to form a kissing-loop structure. This type of structure has been shown to be important in enterovirus replication. The location of the cre has not been identified.
Genome organization and replication
Genome layout:
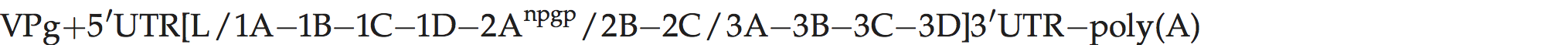
The SVV genome encodes a leader polypeptide of unknown function. It lacks both catalytic residues, present in the aphtho- and erboviruses, which are necessary for proteolytic activity and zinc finger/tyrosine-phosphorylation motifs present in the cardioviruses. 1AB possesses a myristoylation signal at its amino-terminus and is cleaved to VP4 and VP2. VP1 is followed by a short FMDV-like (NPG↓P) 2A.
Antigenic properties
Only a single serotype, Seneca Valley virus 1 (SVV-1), has been recognized.
Biological properties
The only known natural host is the pig but SVV can replicate and cause CPE in a wide range of cell cultures including those derived from pigs, sheep, rabbits, hamsters, monkeys and humans. There is no known association with disease. SVV has potent cytolytic activity and high selectivity for human tumour cell lines having neuroendocrine properties versus adult normal cells. Its use for treatment of human metastatic neuroendocrine cancers has been investigated.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Senecavirus
|
Seneca Valley virus |
|
|
|
Seneca Valley virus 1 [SVV-001] |
[DQ641257] |
(SVV-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Other related viruses which may be members of the genus Senecavirus but have not been approved as species
None reported.
Genus Tremovirus
Type species Avian encephalomyelitis virus
Distinguishing features
Avian encephalomyelitis virus (AEV) is most similar to hepatitis A virus (genus Hepatovirus), but differs by possessing (i) a type IV IRES (HAV is type III), (ii) 2A with a H-box/NC motif and (iii) 2B and 3A polypeptides with little sequence identity to the HAV counterparts.
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
AEV is stable at pH 3.0 and has a buoyant density of 1.31 to 1.33 g cm−3.
Nucleic acid
There is little similarity between the genome sequences of tremoviruses and those of other picornaviruses. The 5′-UTR contains a 5′-terminal hairpin, two putative pseudoknots, and a short (ca. 40 nt) pyrimidine-rich (i.e. not pure poly-C) tract upstream of the type IV IRES. Nucleotide sequence identity between different strains is generally greater than 80%. AEV RNA contains the shortest of all picornavirus 5′-UTRs, at 494 nt. The location of the cre is thought to possibly lie within the 3D region.
Proteins
Similar to hepatoviruses, protein lA of AEV is predicted to be extremely small, does not appear to be myristoylated at its N-terminus, and therefore may not be a component of the mature virus particle.
Genome organization and replication
Genome layout:
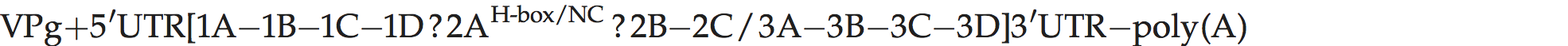
The polyprotein contains only a single protease (3Cpro). There is no L protein, and 2A probably has no proteolytic activity. It is not known if the first primary cleavage occurs between 1D and 2A or between 2A and 2B, nor if one or both of these cleavages are mediated by 3Cpro. The IRES is type IV, similar to avihepatoviruses, sapeloviruses, senecaviruses and teschoviruses. The 2A protein of AEV contains H-box/NC motifs and is distantly related to the 2A of avihepatoviruses, kobuviruses and parechoviruses.
Antigenic properties
Only a single serotype, avian encephalomyelitis virus 1 (AEV-1), has been recognized.
Biological properties
AEV causes encephalomyelitis in young chickens, pheasants, quail and turkeys. It can be transmitted both vertically and by the fecal–oral route; field strains are enterotropic. A live, highly enterotropic AEV vaccine is widely used to control the disease.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Tremovirus
|
Avian encephalomyelitis virus |
|
|
|
Avian encephalomyelitis virus 1 [Calnek] |
[AJ225173] |
(AEV-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Tremovirus but have not been approved as species
None reported.
Genus Avihepatovirus
Type species Duck hepatitis A virus
Virion properties
Morphology
No surface morphology is visible by EM.
Physicochemical and physical properties
DHAV-1 is both heat- and acid-stable.
Nucleic acid
Duck hepatitis A viruses have a 5′ UTR of 625–655 nt which contains a type IV IRES. The location of the cre has not been identified. The 3′ UTR is particularly long, at around 318 nt.
Genome organization and replication
Genome layout:
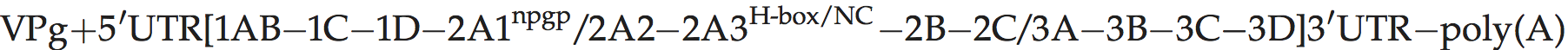
The 1AB polypeptide lacks a myristoylation signal. The genome sequences of all three DHAV types have three 2A motifs: (i) NPGP; (ii) AIG1-like protein containing a GxxGxGKS NTP-binding motif; and (iii) a H-box/NC motif (similar to the 2A of parecho-, kobu- and tremoviruses). However, it is not clear if this genome region encodes one, two or three mature polypeptides.
Antigenic properties
Duck hepatitis A virus is divided into three genetic types: DHAV-1 to -3. DHAV-2 and DHAV-3 are not neutralized by DHAV-1 antiserum; however, the relationship between types 2 and 3 remains unstudied.
Biological properties
DHAV causes a highly fatal contagious disease of young ducklings, 1–28 days of age. The onset of the disease is very rapid, it spreads quickly through the flock and may cause up to 90% mortality. Sick ducklings develop spasmodic contractions of their legs and die within an hour in a typical “arched-backward” position. The liver is enlarged and shows hemorrhagic spots. A DHAV-1 live-attenuated vaccine is widely used.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Avihepatovirus
|
Duck hepatitis A virus |
|
|
|
|
|
Duck hepatitis A virus 1 [R85952] |
[DQ226541] |
(DHAV-1) |
|
|
(Duck hepatitis virus 1) |
|
|
|
|
Duck hepatitis A virus 2 [04G] |
[EF067923] |
(DHAV-2) |
|
|
Duck hepatitis A virus 3 [AP-03337] |
[DQ256132] |
(DHAV-3) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Avihepatovirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Picornaviridae but have not been approved as species
A number of picornaviruses have recently been sequenced, but not formally classified. They include the first confirmed picornaviruses of wild birds, reptiles and fish.
Bat kobu-like virus Nucleotide sequence analysis of about 60% of the genome of a picornavirus detected in bat guano suggests a distant relationship with the kobuviruses.
Bluegill picornavirus Genome layout:

Bluegill picornavirus (BGPV) infects the bluegill (Lepomis macrochirus) freshwater fish in North American lakes. BGPV particles are icosahedral and average 30 nm in diameter. The virus can be grown in a cell line, BF-2, derived from bluegill fry. The genome layout is typical of a picornavirus except that there appears to be at least two 2A polypeptides each having an NPG↓P motif. The first is predicted to be 17 aa analogous to the FMDV 2A, while the second is 136 aa in length. BGPV may represent a novel picornavirus genus. Within the 3′ UTR, there is a poly(C) tract of 21 residues preceding the poly(A) tail.
Carp picornavirus Genome layout:
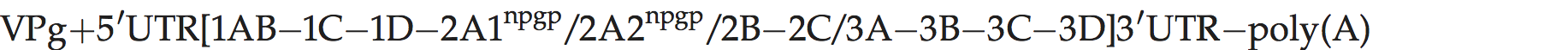
Carp picornavirus (CPV) infects the common carp (Cyprinus carpio), a freshwater fish. 7634 nt of the genome including 524 nt of the 5′ UTR and 284 nt of the unusually long 3′ UTR have been sequenced. Like BGPV, CPV is predicted to possess two 2A polypeptides both with NPG↓P motifs, the second of which is 133 aa in length. CPV may represent a novel picornavirus genus or may belong in the same genus as BGPV.
Eel picornavirus Genome layout:
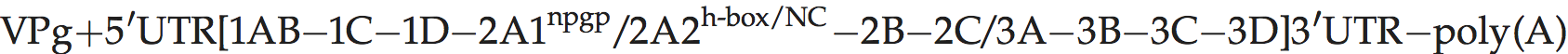
Eel picornavirus (EPV) was isolated from a common eel (Anguilla anguilla) in Lake Constance on the river Rhine. EPV can be propagated in eel kidney 1 (EK-1) cells. It is pathogenic in experimentally infected glass eels and induces a high mortality. 7501 nucleotides of the genome were sequenced. The 3′ UTR comprises 235 nt, part of the 5′ UTR has to be determined. The 3D sequence shows closest similarity to parechoviruses, tremovirus and seal picornavirus. The polyprotein has no leader peptide, a 2A1 protein with NPG↓P motif and a 2A2 protein with the H-box/NC sequence motives. Protein 1AB appears not to be cleaved. EPV may represent a novel picornavirus genus.
Human cosaviruses Genome layout:
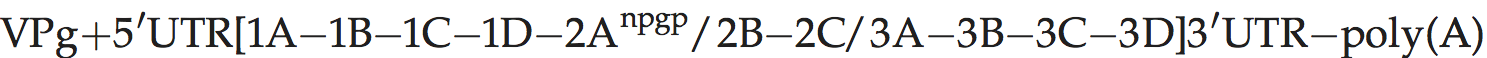
Cosaviruses have been detected in human stools, but have not yet been cultivated. It is proposed that they belong to five different species, based on the same sequence distance criteria used to distinguish human enterovirus species. They are most closely related to cardio- and senecaviruses, but lack a leader polypeptide. The 2A/2B junction is predicted based on a NPG↓P sequence motif, however, evidence for a conserved cleavage site between VP1 and 2A poor. The 5′ UTR is long (>1100 nt) and is predicted to contain a type II IRES. It has been suggested that human cosaviruses may represent a novel picornavirus genus.
Human klassevirus/salivirus Genome layout:
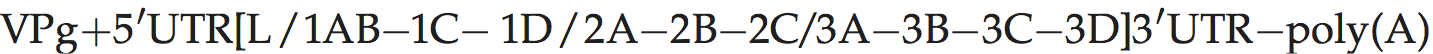
Two very closely related (with a polyprotein aa identity of >95%) viruses have recently been described and named klassevirus (from kobu-like viruses associated with stool and sewage) and salivirus (from stool Aichi-like viruses). The IRES possibly belongs to type II. These viruses possess a leader polypeptide of unknown function. 1AB may not be cleaved to give 1A and 1B, but like kobuviruses 1AB does have a myristoylation signal GxxxT). Kobuviruses possess H-Box/NC motifs in their predicted 2A proteins, however, these are absent in klasse/saliviruses. It has been suggested that the 2A protein is a trypsin-like protease based on the detection of H, C, and D catalytic triad residues at approximately the same location as in the 2A of rhinoviruses and enteroviruses.
Seal picornavirus Genome layout:
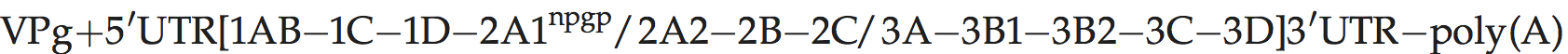
Seal picornavirus 1 (SePV-1) has been isolated from Arctic ringed seals (Pusa hispida) in northern Canada and common (harbour) seals (Phoca vitulina) in the North Sea. Any role in disease is unclear. SePV-1 is predicted to possess two tandemly-repeated VPgs, an uncleaved 1AB (which also lacks a myristoylation motif) and two 2A polypeptides. The IRES is type IV. It has been suggested that SePV-1 may represent a novel picornavirus genus.
Tortoise picornavirus (aka virus “X”) Genome layout:
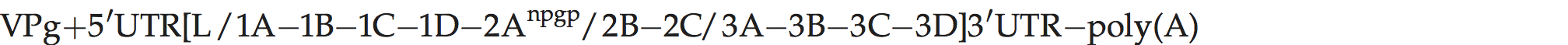
A virus isolated from a spur-thighed tortoise (Testudo graeca) has been cultivated in Terrapene heart cells causing a lytic infection. Approximately 7 kb of the genome has been sequenced (although an unknown length of the 5′ UTR remains to be determined). The predicted polyprotein has a typical picornavirus layout. The short leader polypeptide (52 aa) contains seven cysteine residues and has some similarity to metallothionein-like proteins. The capsid region is most closely related to the erboviruses. The 2B polypeptide is distantly related to that of TMEV, while 2C, 3C and 3D are most closely related to human cosavirus A, FMDV and Aichi virus, respectively. The predicted junction between 2A and 2B is NPG↓P, as it is in the most closely related picornaviruses. The 3′ UTR is 232 nt long and extremely A+T rich (87%). It has been suggested that TPV may represent a novel picornavirus genus.
Turdiviruses: Genome layout:
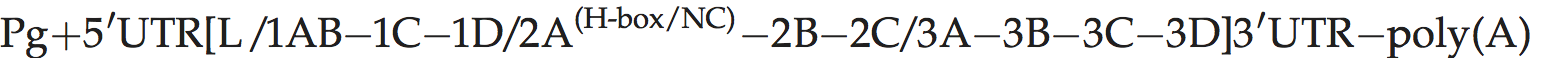
Three novel picornaviruses, named turdiviruses 1, 2 and 3 (TV-1, TV-2 and TV-3), have been identified in birds of different genera in the family Turdidae. Regions P1, P2 and P3 of the three turdiviruses possess, respectively, <40, <40 and <50% amino acid identities with those of other picornaviruses. Moreover, P1, P2 and P3 of TV-1 also possessed, respectively, <40, <40 and <50% amino acid identities with those of TV-2 and TV-3. Phylogenetic analysis revealed that TV-1, TV-2 and TV-3 were distantly related to members of the genus Kobuvirus. The genomic features of TV-2 and TV-3 were also distinct from TV-1, including a lower G+C content and a shorter predicted leader polypeptide. The 2A of TV-1, but not TV-2 or TV-3, appears to be of the H-box/NC type. It has been suggested that the turdiviruses may fall into two novel picornavirus genera.
Turkey hepatitis virus: Genome layout:
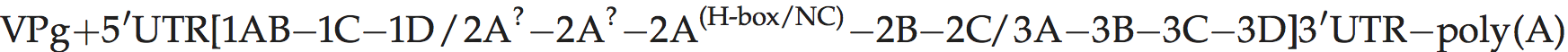
Turkey hepatitis virus (THV) has been identified in the liver, bile, intestine, serum and cloacal swabs of poults with disease, but not in clinically normal birds. The genome sequence has been determined for two related viruses and each was over 9 kb, the longest amongst the picornaviruses. The 5′ UTR IRES shares some similarity with that of DHAV and therefore may be of type IV. Comparative predictions suggest that 1AB (VP0) is neither cleaved nor myristoylated. There is a large insertion of about 1.2 kb between VP1 and a recognizable 2A-like region containing Hbox-NC motifs (also present in hepato-, kobu-, parecho- and tremoviruses). Prediction of cleavage sites within this region suggests that an additional two distinct 2A polypeptides of unknown function may be present. THV is most closely related to the kobuviruses and the unclassified human klasse/saliviruses and avian turdiviruses. The presence of a poly(A) tail is presumed, but not yet proven.
|
Bat kobu-like virus |
[HM228880 to HM228884] |
(BKLV) |
|
Bluegill picornavirus 1 [04-032] |
|
(BGPV-1) |
|
Carp picornavirus 1 [F37/06] |
|
(CPV-1) |
|
Eel picornavirus 1 [F15/05] |
|
(EPV-1) |
|
Human cosavirus A1 [0553] |
[FJ438902] |
(HCoSV-A1) |
|
Human cosavirus A2 [6344] |
[FJ438903] |
(HCoSV-A2) |
|
Human cosavirus A3 [6572] |
[FJ438904] |
(HCoSV-A3) |
|
Human cosavirus A4 [5006] |
[FJ438905, FJ438906] |
(HCoSV-A4) |
|
Human cosavirus B1 [2263] |
[FJ438907] |
(HCoSV-B1) |
|
Human cosavirus C1 [5152] |
[FJ442995] |
(HCoSV-C1) |
|
Human cosavirus D1 [5004] |
[FJ438908] |
(HCoSV-D1) |
|
Human cosavirus E1 [Australia/81] |
[FJ555055] |
(HCoSV-E1) |
|
Human klassevirus 1 [02394-01] |
[GQ184145] |
(HKV-1) |
|
Salivirus [NG-J1 ] |
[GQ179640] |
(SaV) |
|
Seal picornavirus 1 [HO.02.21] |
[EU142040] |
(SePV-1) |
|
Sheep picornavirus 1 [VS65.60] |
|
(ShPV-1) |
|
Tortoise picornavirus 1 (virus X) [TGT1A/96] |
|
(TPV-1) |
|
Turdivirus 1 [00356 & 00805] |
[GU182406, GU182407] |
(TV-1) |
|
Turdivirus 2 [10717 & 007167] |
[GU182408, GU182409] |
(TV-2) |
|
Turdivirus 3 [10878 & 00742] |
[GU182410, GU182411] |
(TV-3) |
|
Turkey hepatitis virus [2993D & 0091.1] |
[HM751199, HQ189775] |
(THV) |
Names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
A number of other candidate picornaviruses exist for which no sequence data are yet available. These have been described as picornaviruses or picorna-like viruses based mainly on EM morphology and size.
|
Aesculapian snake picorna-like virus |
(ASPLV) |
|
Avian entero-like virus 2 [EF84/700] |
(AELV-2) |
|
Barramundi virus 1 |
(BaV) |
|
Boa constrictor picorna-like virus |
(BCPLV) |
|
Cockatoo entero-like virus |
(CELV) |
|
European smelt picornavirus |
(ESV) |
|
Grass carp picornavirus |
(GCPV) |
|
Greasy grouper virus |
(GGV) |
|
Guineafowl transmissible enteritis virus |
(GTEV) |
|
Juruaca virus [BeAn 401933] |
(JURV) |
|
Malabar grouper virus |
(MGV) |
|
Rainbow smelt picornavirus |
(RSPV) |
|
Salmonid viruses |
|
|
Sandbar shiner virus |
(SSV) |
|
Sea-bass virus 1 |
(SBV-1) |
|
Sikhote-Alyn virus [LEIV 113P] |
(SAV) |
|
Smelt virus 1 |
(SmV-1) |
|
Smelt virus 2 |
(SmV-2) |
|
Syr-Darya Valley fever virus [Kaz-3] |
(SDFV) |
|
Turbot virus 1 |
(TuV-1) |
|
Turkey entero-like virus |
(TELV) |
|
Turkey pseudo enterovirus 1 |
(TPEV-1) |
|
Turkey pseudo enterovirus 2 |
(TPEV-2) |
Phylogenetic relationships within the family
Viruses in each genus are phylogenetically distinct from members of other genera in those genome regions which are homologous, i.e. P1cap, 2Chel, 3Cpro and 3Dpol.
Phylogenetic trees showing the relationships between the genera, species and unclassified members of the family Picornaviridae: (A.P1) protein P1; (B.2C) protein 2C; and (C.3CD) proteins 3C+3D. The neighbor-joining trees were produced using MEGA 4.0 and the poisson model for correction of multiple substitutions. Abbreviations: AEV, Avian encephalomyelitis virus; AiV, Aichi virus; ASV, Avian sapelovirus; BEV, Bovine enterovirus; BGPV, bluegill picornavirus; BKV, Bovine kobuvirus; BRAV, bovine rhinitis A virus; BRBV, Bovine rhinitis B virus; CPV, carp picornavirus; DHAV, Duck hepatitis A virus; EMCV, Encephalomyocarditis virus; EPV, eel picornavirus; ERAV, Equine rhinitis A virus; ERBV, Equine rhinitis B virus; FMDV, Foot-and-mouth disease virus; HAV, Hepatitis A virus; HCoSV, human cosavirus; HEV, human enterovirus; HKV, human klassevirus; HPeV, Human parechovirus; HRV, human rhinovirus; LV, Ljungan virus; PEV-B, Porcine enterovirus B; PKV, porcine kobuvirus; PSV, Porcine sapelovirus; PTV, Porcine teschovirus; SePV, seal picornavirus; SEV-A, Simian enterovirus A; SSV, Simian sapelovirus; SVV, Seneca Valley virus; ThV, Theilovirus; THV, turkey hepatitis virus; TPV, tortoise picornavirus; TV, turdivirus.
Similarity with other taxa
The family Picornaviridae together with the families Dicistroviridae, Iflaviridae, Marnaviridae and Secoviridae form the order Picornavirales. There are also similarities to the families Caliciviridae and Potyviridae, including the order of the Hel-VPg-Pro-Pol non-structural polypeptides.
Derivation of names
Aphtho: from Greek aphthae, “vesicles in the mouth”; English: aphtha, “thrush”; French: fièvre aphteuse.
Avihepato: from avian and Greek hepatos, “liver”.
Cardio: from Greek kardia, “heart”.
Entero: from Greek enteron, “intestine”.
Erbo: for equine rhinitis B virus.
Hepato: from Greek hepatos, “liver”.
Kobu: from Japanese kobu, “knuckle” (reference to surface structure of virus particle).
Parecho: from par(a)echo (echo, the former name of the type species, an acronym for “enteric cytopathic human orphan”).
Picorna: from the prefix “pico” (= “micro-micro”) and RNA.
Sapelo: from simian, avian and porcine entero-like viruses.
Seneca: from Seneca Valley virus.
Tescho: from Teschen disease.
Tremo: from an alternative name for avian encephalomyelitis, epidemic tremor.
Further reading
Journals and books
2011. In: Ehrenfeld, E., Domingo, E., Roos, R.P., The Picornaviruses. ASM Press, Washington, DC.
Ferrer-Orta, C., Arias, A., Escarmís, C., Verdaguer, N., 2006. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16, 27–34.
Lin, J.Y., Chen, T.C., Weng, K.F., Chang, S.C., Chen, L.L., Shih, S.R., 2009. Viral and host proteins involved in picornavirus life cycle. J. Biomed. Sci. 16, 103.
Lukashev, A.N., 2010. Recombination among picornaviruses. Rev. Med. Virol. 20, 327–333.
Pacheco and Martinez-Salas, 2010 Pacheco, A. Martinez-Salas, E. (2010). Insights into the biology of IRES elements through riboproteomic approaches. J. Biomed. Biotechnol., Volume 2010, Article ID 458927 12pp.
Wang, H.M., Liang, P.H., 2010. Picornaviral 3C protease inhibitors and the dual 3C protease/coronaviral 3C-like protease inhibitors. Expert Opin. Ther. Pat. 20, 59–71.
Witwer, C., Rauscher, S., Hofacker, I.L., Stadler, P.F., 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29, 5079–5089.
Websites
The Picornaviruses Pages: www.picornaviridae.com/
Picornaviridae Study Group: www.picornastudygroup.com/
Contributed by
Knowles, N.J., Hovi, T, Hyypiä, T., King, A.M.Q., Lindberg, A.M., Pallansch, M.A., Palmenberg, A.C., Simmonds, P., Skern, T., Stanway, G., Yamashita, T. and Zell, R.
Figures
Figure 1 Pictures of picornavirus structures; (a) poliovirus 1 (2PLV); (b) Mengo (2MEV); (c) TMEV (1TME); (d) FMDV O (1FOD); (e) SVV (3CJI); the bar represents 10 nm (images courtesy of Jean-Yves Sgro, with permission). (f) Diagram of a picornavirus particle. The surface shows proteins VP1, VP2 and VP3. The fourth capsid protein, VP4, is located about the internal surface of the pentameric apex of the icosahedron. (g) Negative contrast electron micrograph of poliovirus (PV) particles; the bar represents 100 nm (courtesy of Ann C. Palmenberg). (h) Negative contrast electron micrograph of Aichi virus (genus Kobuvirus) showing surface structure; the bar represents 50 nm
(courtesy of Teruo Yamashita).

Figure 2 Percentage G+C content of picornavirus genomes.
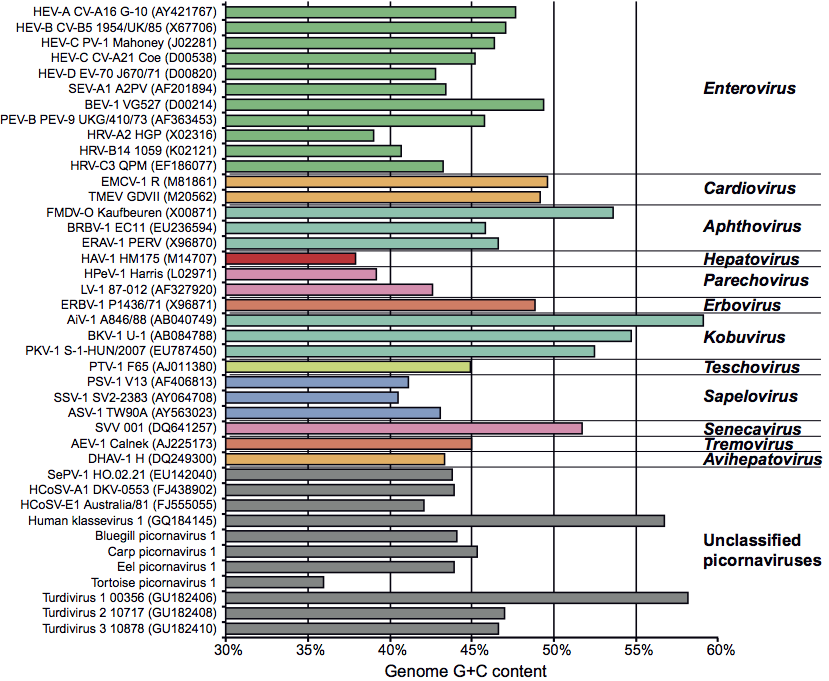
Figure 3 Genome structure and gene organization of members of the family Picornaviridae. Each of the 12 genera is represented, as are species where there is a significant difference within a genus. Circles within the 5 UTR indicate poly(C) tracts that are present in some members. The 1A gene products of many members are myristylated at the amino terminal glycine. The 5 UTR is followed by a long ORF encoding the polyprotein, that is in turn followed by the 3 UTR and a poly(A) tail. The eventual cleavage products of the polyprotein are indicated by vertical lines and different shading. The nomenclature of the polypeptides follows an L:4:3:4 scheme corresponding to the genes (numbers) encoded by the L, P1, P2, P3 regions. The P1 region encodes the structural proteins 1A, 1B, 1C and 1D, also referred to as VP4, VP2, VP3 and VP1, respectively. VP0 (1AB) is the intermediate precursor for VP4 and VP2 and in avihepato-, kobu- and parechoviruses it remains uncleaved. In all viruses 3C is a protease, in enteroviruses 2A is a protease, while in all viruses 3D is considered to be a component of the RNA replicase. Only foot-and-mouth disease virus encodes 3 VPg proteins that map in tandem.
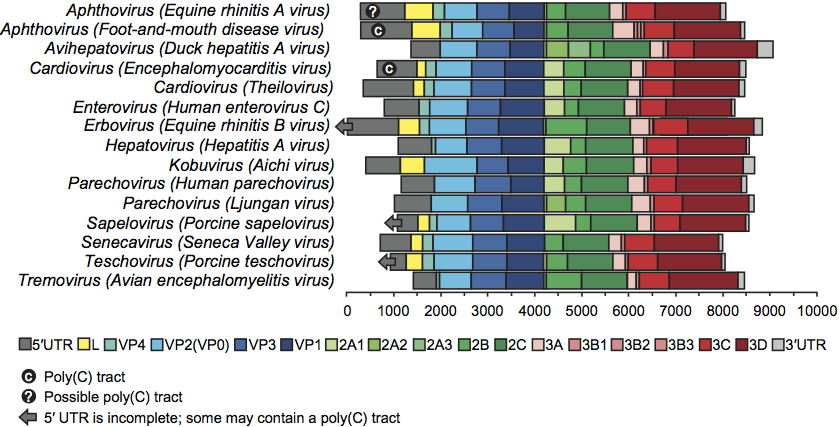
Figure 4 Phylogenetic trees showing the relationships between the genera, species and unclassified members of the family Picornaviridae: (A.P1) protein P1; (B.2C) protein 2C; and (C.3CD) proteins 3C+3D. The neighbor-joining trees were produced using MEGA 4.0 and the poisson model for correction of multiple substitutions. Abbreviations: AEV, Avian encephalomyelitis virus; AiV, Aichi virus; ASV, Avian sapelovirus; BEV, Bovine enterovirus; BGPV, bluegill picornavirus; BKV, Bovine kobuvirus; BRAV, bovine rhinitis A virus; BRBV, Bovine rhinitis B virus; CPV, carp picornavirus; DHAV, Duck hepatitis A virus; EMCV, Encephalomyocarditis virus; EPV, eel picornavirus; ERAV, Equine rhinitis A virus; ERBV, Equine rhinitis B virus; FMDV, Foot-and-mouth disease virus; HAV, Hepatitis A virus; HCoSV, human cosavirus; HEV, human enterovirus; HKV, human klassevirus; HPeV, Human parechovirus; HRV, human rhinovirus; LV, Ljungan virus; PEV-B, Porcine enterovirus B; PKV, porcine kobuvirus; PSV, Porcine sapelovirus; PTV, Porcine teschovirus; SePV, seal picornavirus; SEV-A, Simian enterovirus A; SSV, Simian sapelovirus; SVV, Seneca Valley virus; ThV, Theilovirus; THV, turkey hepatitis virus; TPV, tortoise picornavirus; TV, turdivirus.