Family: Marnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Marnavirus
Type species Heterosigma akashiwo RNA virus
Virion properties
Morphology
At present, the only characterized representative of the family Marnaviridae is Heterosigma akashiwo RNA virus (HaRNAV). Based on electron micrographs, HaRNAV virions are approximately 25 nm in diameter, polyhedral in shape, do not appear to have an envelope, and have no discernible projections (Figure 1).
Physicochemical and physical properties
HaRNAV is not sensitive to chloroform.
Nucleic acid
HaRNAV has an 8.6 kb ssRNA genome containing a single ORF. The genome has a poly(A) tail at the 3′ terminus. The 5′- and 3′-UTRs are 483 and 361 nt long, respectively, accounting for a total of 9.8% of the genome. Computer predictions (mfold 3.0) of secondary structure of the 5′-UTR and a notable pyrimidine-rich stretch of sequence upstream of the predicted start codon, suggest the presence of an IRES, a feature observed in many picorna-like viruses. The genome sequence has two large pseudo repeats in the 5′- and 3′-UTRs. A 136 nt sequence in the 5′-UTR shares 123 exact bases with a 137 nt sequence in the 3′ end. These repeated sequences may have some function in replication and/or translation.
Proteins
The major structural proteins of HaRNAV are characterized in Table 1. The 33 and 29 kDa protein sequence revealed similarities to the VP3 proteins from members of the families Dicistroviridae and Picornaviridae and the VP1 proteins from the Dicistroviridae, respectively. Cleavage sites delineating the structural protein domains lack a recognizable pattern raising the possibility that there may be more than one protease involved in polyprotein processing.
Table 1 Characteristics of major HaRNAV structural proteins
| Protein (kDa)a | Position of N-teminus in polyproteinb | Putative sequence at cleavage siteb | Location on genome map (Figure 2) |
| 39 | 1990 | PTST-SEIV | 3 |
| 33 | 2318 | FVST-SEII | 5 |
| 29 | 2060 | LFGY-SRPP | 4 |
| 26 | 1776 | EKLL-TETL | 1 |
| 24 | 1810 | RPGE-VDGD | 2 |
a Based on SDS-PAGE.
b Based on N-terminal sequencing and genome sequence analysis.
Lipids
Undetermined.
Carbohydrates
Undetermined.
Genome organization and replication
The map of protein domains within the predicted HaRNAV polyprotein sequence is shown in Figure 2. Domains were identified on the basis of similarities with conserved domains for other members of the order Picornavirales including helicases, RdRps, and CPs. HaRNAV also encodes an amino acid sequence that resembles the 3C cysteine proteinases of members of the family Picornaviridae and that is also related to the chymotrypsin-related serine protease catalytic domain. A VPg-like protein, characteristic of most picorna-like viruses, has not been identified.
Antigenic properties
Undetermined.
Biological properties
The host range of HaRNAV is restricted to specific strains of Heterosigma akashiwo. Of 15 host strains isolated from the Northwest Pacific, Western Atlantic Ocean and Japanese coastal waters, five were permissive to HaRNAV infection. HaRNAV replication appears to be cytolytic. Cytopathic effects begin approximately 48 h after infection. Ultrastructural changes include swelling of the endoplasmic reticulum, vacuolation and disintegration of the cytoplasm, and the appearance of fibrous material in vacuolated areas. Particles of HaRNAV are distributed in the cytoplasm in crystalline arrays or as individuals.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Marnavirus
| Heterosigma akashiwo RNA virus |
|
|
| Heterosigma akashiwo RNA virus SOG263 | [AY337486] | (HaRNAV-SOG263) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Marnavirus but have not been approved as species
Closely related viruses have been identified in marine environments, although these were not isolated and studied directly. A degenerate RT-PCR approach targeting the RdRp gene found a number of related RdRp sequences that were different from the genome-sequenced strain of HaRNAV, SOG263. These RdRp sequences did have a high level of identity with HaRNAV and likely represented different strains of the virus.
List of unassigned species in the family Marnaviridae
None.
List of other related viruses which may be members of the family Marnaviridae but have not been approved as species
| Chaetoceros socialis f. radians RNA virus | [AB469874] | (CsfrRNAV) |
| Chaetoceros tenuissimus RNA virus | [AB375474] | (CtenRNAV) |
| Rhizosolenia setigera RNA virus | [AB243297] | (RsRNAV) |
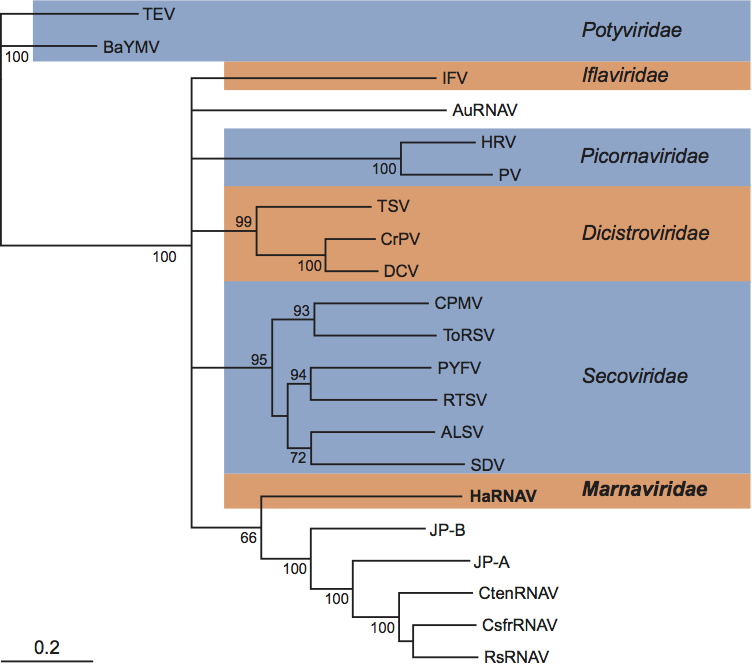
Phylogenetic relationships within the family
Not applicable.
Similarity with other taxa
The HaRNAV genome is composed of one molecule of positive sense ssRNA that exhibits the 2C-3Cpro-3Dpol gene order, and the particles are icosahedral with a diameter of about 25 nm, criteria that are consistent with placing the family Marnaviridae within the order Picornavirales. However, the structure of the viral genome and the patterns of sequence relationships of HaRNAV proteins to those of other viruses within the Picornavirales clearly show that HaRNAV does not belong within any of the other established families. The HaRNAV genome structure is most like the potyviruses (e.g., tobacco etch virus) in that the non-structural protein domains are located at the N-terminus and the structural proteins are at the C-terminus in a single large polyprotein encoded on a mono-partite genome. However, potyvirus capsids are filamentous and phylogenetic analyses demonstrated no significant relationship with this family (Figure 3). Moreover, a phylogenetic analysis of picorna-like RdRps does not place the HaRNAV sequence within any established family of picorna-like viruses (Figure 3).
HaRNAV appears to be most closely related to three unclassified viruses that infect species of marine diatoms, Chaetoceros socialis f. radians RNA virus (CsfrRNAV), Chaetoceros tenuissimus RNA virus (CtenRNAV) and Rhizosolenia setigera RNA virus (RsRNAV). HaRNAV, CsfrRNAV, CtenRNAV and RsRNAV have several characteristics in common including icosahedral morphology, a positive sense single stranded genome with a poly(A) tail, the non-structural ORF gene order, the organization of the non-structural protein domains relative to the structural proteins, and all infect protistan hosts. However, whereas the three diatom virus genomes are dicistronic, the HaRNAV genome encodes a single polyprotein. Phylogenetic analysis based on an alignment of the RdRp suggests that HaRNAV clusters with, but is distantly related to, CsfrRNAV, CtenRNAV and RsRNAV (Figure 3). There is a proposal under consideration to create a new genus, named Bacillarnavirus, for these three diatom viruses. The genus would be unassigned in the order Picornavirales.
Metagenomic libraries and single-gene surveys targeting marine RNA viruses have uncovered a diverse group of RNA virus phylotypes that appear to be from viruses related to Picornavirales. The genomes JP-A and JP-B were assembled from reverse-transcribed whole-genome shotgun libraries constructed from virus communities harvested from the Strait of Georgia, British Columbia. The genomes of JP-A and JP-B are 9.2 and 8.8 kb in length, respectively, have poly(A) tails, are dicistronic, and have a similar organization to the genomes of RsRNAV, CsfrRNAV and CtenRNAV. JP-A and JP-B appear to be most closely related to the diatom viruses discussed above, nevertheless, they belong to a clade that includes HaRNAV (Figure 3). Phylogenetic analysis shows that the vast majority of RNA virus RdRp sequences amplified from marine virus communities form a well-supported clade with homologous sequences from the Picornavirales that infect marine protists, including HaRNAV. However, with the exception of the nearly identical HaRNAV-like sequences discussed above, these environmental viral phylotypes appear to be most closely related to CsfrRNAV, CtenRNAV and RsRNAV. In general, the taxonomy of Picornavirales reflects the taxonomy of their hosts, therefore it is likely that close relatives of HaRNAV are absent because an RNA virus infecting a close relative of Heterosigma akashiwo has yet to be isolated. Nevertheless, the diverse number of RNA virus phylotypes with unknown hosts recovered from a limited number of samples intimates that the discovery of new members of the Marnaviridae is just a matter of time.
Derivation of name
Marna: from Latin mare, “sea”, and RNA.
Further reading
Culley et al., 2006 A.I. Culley, A.S. Lang, C.A. Suttle, Metagenomic analysis of coastal RNA virus communities. Science. 312 (2006) 1795–1798.
Culley et al., 2007 A.I. Culley, A.S. Lang, C.A. Suttle, The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol. J. 4 (2007) 69.
Culley and Steward, 2007 A.I. Culley, G.F. Steward, New genera of RNA viruses in subtropical seawater inferred from polymerase gene sequences. Appl. Environ. Microbiol. 73 (2007) 5937–5944.
Lang et al., 2003 A.S. Lang, A.I. Culley, C.A. Suttle, Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology. 310 (2003) 359–371.
Lang et al., 2009 A.S. Lang, M.L. Rise, A.I. Culley, G.F. Steward, RNA viruses in the sea. FEMS Microbiol. Rev. 33 (2009) 295–323.
Nagasaki, 2008 K. Nagasaki, Dinoflagellates, diatoms and their viruses. J. Microbiol. 46 (2008) 235–243.
Nagasaki et al., 2004 K. Nagasaki, Y. Tomaru, N. Katanozaka, Y. Shirai, K. Nishida, S. Itakura, M. Yamaguchi, Isolation and characterization of a novel single-stranded RNA virus infecting the bloom-forming diatom Rhizosolenia setigera. Appl. Environ. Microbiol. 70 (2004) 704–711.
Shirai et al., 2006 Y. Shirai, Y. Takao, H. Mizumoto, Y. Tomaru, D. Honda, K. Nagasaki, Genomic and phylogenetic analysis of a single-stranded RNA virus infecting Rhizosolenia setigera (Stramenopiles: Bacillariophyceae). J. Mar. Biol. Ass. U.K. 86 (2006) 475–483.
Shirai et al., 2008 Y. Shirai, Y. Tomaru, Y. Takao, H. Suzuki, T. Nagumo, K. Nagasaki, Isolation and characterization of a single-stranded RNA virus infecting the marine planktonic diatom Chaetoceros tenuissimus Meunier. Appl. Environ. Microbiol. 74 (2008) 4022–4027.
Tai et al., 2003 V. Tai, J.E. Lawrence, A.S. Lang, A.M. Chan, A.I. Culley, C.A. Suttle, Characterization of HaRNAV, a single-stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae). J. Phycol. 39 (2003) 343–352.
Tomaru et al., 2009 Y. Tomaru, Y. Takao, H. Suzuki, T. Nagumo, K. Nagasaki, Isolation and characterization of a single-stranded RNA virus infecting the bloom-forming diatom Chaetoceros socialis. Appl. Environ. Microbiol. 75 (2009) 2375–2381.
Contributed by
Culley, A.I., Lang, A.S. and Suttle, C.A.
Figures
Figure 1 (Left) Diagrammatic representation of the possible structure of Heterosigma akashiwo RNA virus (HaRNAV) particles. (Right) Electron micrograph of HaRNAV particles stained with phosphotungstic acid. The bar represents 50 nm.
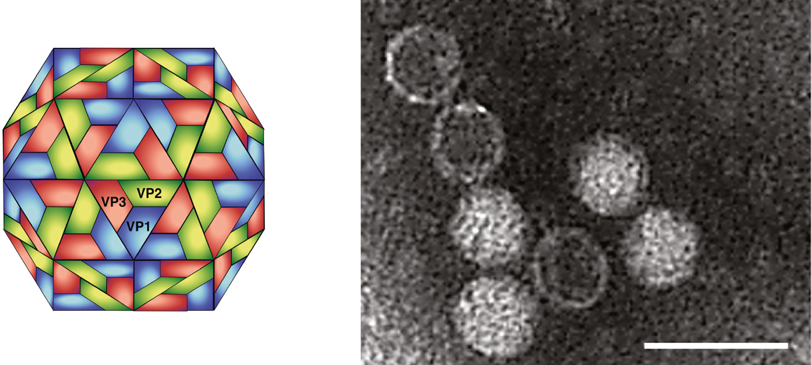
Figure 2 Representation of the genome organization of HaRNAV. The location of conserved picorna-like protein domains are indicated within the polyprotein box: H, helicase; P, protease; RdRp, RNA-dependent RNA polymerase; VP2, VP3 and VP1, structural proteins. The locations of N-termini found by sequencing the HaRNAV structural proteins are shown by black lines in the box and numbered to indicate the sequences at these processing sites, given in Table 1.
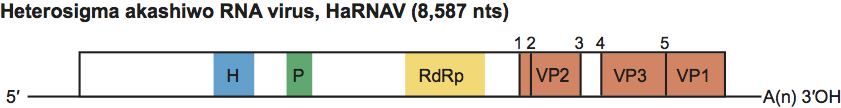
Figure 3 Bayesian maximum likelihood phylogenetic tree of RdRp sequences from the order Picornavirales. Residues 13621619 of the HaRNAV polyprotein that represent the conserved regions I-VIII and the corresponding regions from the other viruses included were aligned with DIALIGN-TX. Amino acid alignments were transformed into likelihood distances with Mr Bayes v3.1.2 using 1,100,000 generations. Bayesian clade credibility values are shown for relevant nodes. The Bayesian scale bar indicates a distance of 0.2. Taxonomic groups are indicated where known. TEV, tobacco etch virus; BaYMV, barley yellow mosaic virus; IFV, infectious flacherie virus; AuRNAV, Aurantiochytrium single stranded RNA virus; HRV, human rhinovirus; PV, poliovirus; TSV, taura syndrome virus; CrPV, cricket paralysis virus; DCV, Drosophila C virus; CPMV, cowpea mosaic virus; ToRSV, tomato ringspot virus; PYFV, parsnip yellow fleck virus; RTSV, rice tungro spherical virus; ALSV, apple latent spherical virus; SDV, satsuma dwarf virus; CtenRNAV, Chaetoceros tenuissimus RNA virus; CsfrRNAV, Chaetoceros socialis f. radians RNA virus; RsetRNAV, Rhizosolenia setigera RNA virus. JP-A and JP-B are environmental sequences.