Family: Iflaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Iflavirus
Type species Infectious flacherie virus
Virion properties
Morphology
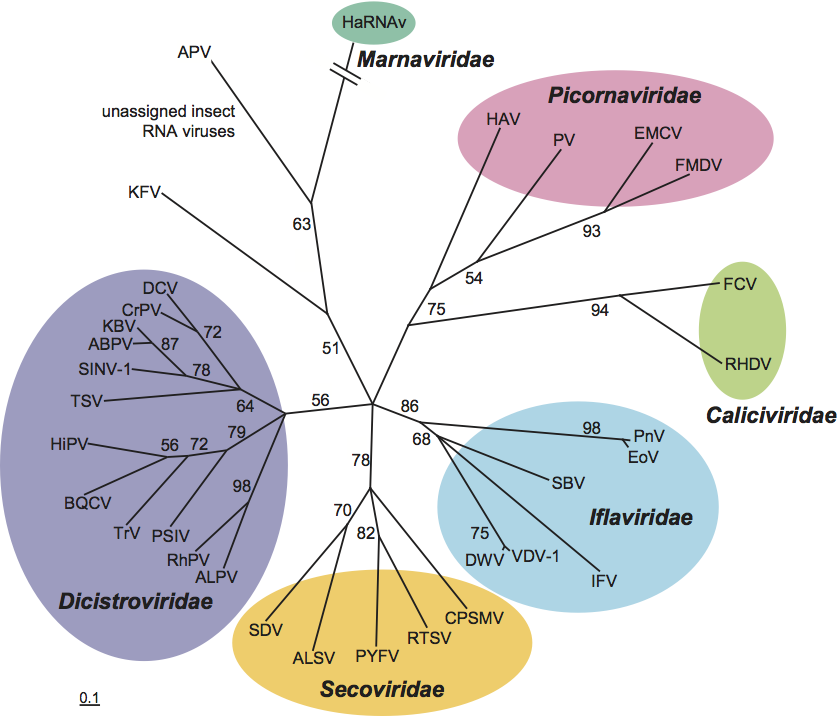
Virions are roughly spherical and exhibit icosahedral symmetry with a diameter of 26–30 nm. Virions have no envelope and no distinctive surface structures (Figure 1).
Physicochemical and physical properties
Virions have a buoyant density of between 1.33 and 1.38 g cm−3.
Nucleic acid
Virions contain one molecule of linear, positive sense, single stranded RNA. The genome has a size ranging from 8800–10,100 nt and contains a single large ORF encoding a polyprotein of 2858–3085 amino acids. A small genome-linked virus protein, VPg is linked covalently to the 5′ end of the genomic RNA. The untranslated regions (UTRs) flanking both ends of the ORF vary in size by species. The 5′-UTR of infectious flacherie virus (IFV) and sacbrood virus (SBV) (130–180 nt) is significantly shorter than that of the other iflaviruses (400–1200 nt).
Proteins
All proteins arise by proteolytic cleavage of a single polyprotein. Mature virions contain three major structural proteins (VP1, VP2 and VP3) generally between 28 and 44 kDa. A fourth smaller capsid protein (VP4) of around 4–12 kDa has been reported in some species and is located in the second position of the capsid precursor coding region. Minor quantities of the uncleaved precursors have been reported in some species.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome is monopartite and contains a large ORF with genes encoding capsid proteins at the 5′ end and genes encoding the non-structural proteins at the 3′ end of the genome. The capsid proteins, arranged in the order of VP2-VP4-VP3-VP1, are preceded by a short leader protein (L) that is removed from VP2 before capsid assembly and has no known function. VP4 is analogous to VP4 present in some dicistroviruses and in the case of IFV is present as a minor structural component of the capsid. The non-structural proteins include a RNA helicase, a 3C-like cysteine protease, and a RNA-dependent RNA polymerase (5′ to 3′ orientation as illustrated in Figure 2). It is unclear if there are any conserved RNA secondary structures in the UTRs.
The viral RNA is infectious and serves as both genomic and viral mRNA. The replication of viruses occurs in the cytoplasm of infected cells but the mechanism of viral genomic RNA entry into the host cell cytoplasm is unknown. The genomic RNA is translated into a polyprotein which is autocatalytically cleaved into structural and non-structural component peptides. For three members in the family (Ectropis obliqua virus (EoV), Perina nuda virus (PnV) and Varroa destructor virus-1 (VDV-1)), there is evidence to suggest that translation of the polyprotein is mediated by an IRES. Mechanisms of polyprotein processing and the effects on host cell macromolecular synthesis during infection have not been well studied for the members of this family.
Antigenic properties
Honeybee viruses in the genus are serologically distinct. There are no known serological relationships between other members of the genus.
Biological properties
All member viruses have been isolated from arthropod species and appear to have restricted host ranges. IFV is known from the lepidopteran species Bombyx mori and Glyphodes pyloalis. PnV is known only from the lepidopteran Perina nuda. EoV infects the lepidopteran Ectropis obliqua. Deformed wing virus (DWV) and SBV are common viruses of the honey bee, Apis mellifera. VDV-1 was isolated from the honey bee parasitic mite, Varroa destructor. SBV and DWV are distributed globally likely from intensive international trade and transportation of bees and bee products. With the exception of PnV, which replicates in a cell line established from Perina nuda, there are no cell culture systems available for the propagation of other viruses in the group.
Species demarcation criteria in the genus
The species demarcation criteria are:
- Natural host range: species can be differentiated on the basis of their natural host range.
- Sequence identity at the amino acid level between the CPs of isolates and strains of a species is above 90%.
List of species in the genus Iflavirus
| Deformed wing virus |
|
|
| Deformed wing virus-Italy | [AJ489744] | (DWV-IT) |
| Kakugo virus | [AB070959] |
|
| Ectropis obliqua virus |
|
|
| (Ectropis obliqua picorna-like virus) |
|
|
| Ectropis obliqua virus | [AY365064] | (EoV) |
| Infectious flacherie virus |
|
|
| Infectious flacherie virus | [AB000906] | (IFV) |
| Perina nuda virus |
|
|
| (Perina nuda picorna-like virus) |
|
|
| Perina nuda virus | [AF323747] | (PnV) |
| Sacbrood virus |
|
|
| Sacbrood virus-Rothamsted | [AF092924] | (SBV-Roth) |
| Varroa destructor virus-1 |
|
|
| Varroa destructor virus-1-Netherlands | [AY251269] | (VDV-1-NL) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Iflavirus but have not been approved as species
| Bee slow paralysis virus | [EU035616] | (BsPV) |
| Brevicoryne brassicae virus 1 | [EF517277] | (BrBV-1) |
Phylogenetic relationships within the family
Phylogenetic analysis of the complete genomes and deduced amino acid sequence of capsid protein precursors indicate close genetic relationships between DWV and VDV-1 (nucleotide identities 79–89% and amino acid identities 89–98% in different regions of the genomes), reflecting a short evolutionary distance after the separation from a common ancestor. EoV and PnV also show a close genetic relatedness to each other. By contrast, the much larger genetic distances dividing these two pairs of species from each other, and from SBV and IFV, are more typical of the distances between different genera that are seen in some members of the order Picornavirales, e.g. the Picornaviridae and Secoviridae (Figure 3).
Similarity with other taxa
The viruses in this family have a structure and genome arrangement similar to the viruses in the Dicistroviridae, Picornaviridae, Marnaviridae and Secoviridae. The RNA genome contains a single large ORF encoding the capsid proteins at the 5′ end and the non-structural proteins at the 3′ end. The 5′ end of the genome is covalently linked to a small peptide, VPg, which plays an important role in RNA replication. The replicases of iflaviruses resemble those of the dicistroviruses, picornaviruses, marnaviruses and secoviruses by containing sequence motifs typical of RNA helicase (Hel), a chymotrypsin-like 3C protease (Pro), and an RNA-dependent RNA polymerase (RdRp) in the 5′ to 3′ orientation. The RdRp domains are phylogenetically related (Figure 3). The virions are icosahedral with pseudo-T=3 symmetry and a diameter about 30 nm.
Derivation of name
Ifla: from the type species of the genus, Infectious flacherie virus.
Further reading
de Miranda and Genersch, 2010 J.R. de Miranda, E. Genersch, Deformed wing virus. J. Invertebr. Pathol. 103 (Suppl. 1.) (2010) s48–s61.
Gall et al., 2008 O.L. Gall, P. Christian, C.M. Fauquet, A.M.Q. King, N.J. Knowles, N. Nakashima, G. Stanway, A.E. Gorbalenya, Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch. Virol. 153 (2008) 715–727.
Contributed by
Chen, Y.P., Nakashima, N., Christian, P.D., Bakonyi, T., Bonning, B.C., Valles, S.M. and Lightner, D.
Figures
Figure 1 (Left) The outer surface view of the virion of infectious flacherie virus (IFV) along a five-fold axis reconstructed by cryo-electron microscopy. The bar represents 10 nm (courtesy of J. Hong). (Right) Negative contrast electron micrograph of isometric particles of an isolate of IFV. The bar represents 100 nm
(courtesy of H. Bando).
Figure 2 Genome structure of infectious flacherie virus (IFV). The genome encodes a single polyprotein that is autocatalytically cleaved into three major structural proteins (VP2, VP3 and VP1) and the non-structural proteins. The 5 end of the genome carries a covalently linked protein, VPg, which plays an important role in RNA replication and the 3 end of the genome is polyadenylated. The structural proteins are encoded at the 5 end of the polyprotein and the non-structural proteins at the 3 end. The capsid proteins, arranged in the order VP2-VP4-VP3-VP1, are preceded by a short leader protein (L). The approximate positions of the helicase (Hel), protease (Pro) and RNA-dependent RNA polymerase (RdRp) domains in the non-structural protein are shown.
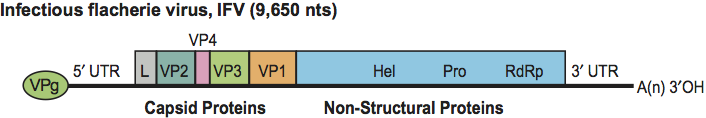
Figure 3 Unrooted phenogram derived from the RdRp domain of the viral non-structural proteins showing the relationships of viruses in the family Iflaviridae and other families of the order Picornavirales (Picornaviridae, Dicistroviridae, Marnaviridae and Secoviridae) and in the closely related family Caliciviridae. The branch length for HaRNAv is shortened to one-fourth because of the distant relationship of this virus. Taxa used (with virus name (abbr.) and accession number) were as follows. Dicistroviridae: acute bee paralysis virus (ABPV) AF150629, aphid lethal paralysis virus (ALPV) AF536531, black queen cell virus (BQCV) AF183905, cricket paralysis virus (CrPV) AF218039, Drosophila C virus (DCV) AF014388, Himetobi P virus (HiPV) AB017037, Kashmir bee virus (KBV) AY27571O, Plautia stali intestine virus (PSIV) AB006531, Rhopalosiphum padi virus (RhPV) AF022937, Solenopsis invicta virus-l (SINV-l) AY6343 14, Taura syndrome virus (TSV) AF277675, Triatoma virus (TrV) AFl78440. Iflaviridae: deformed wing virus (DWV) AY292384, ectropis obliqua virus (EoV) AY365064, infectious flacherie virus (IFV) AB000906, perina nuda virus (PnV) AF323747, sacbrood virus (SBV) AF092924, Varroa destructor virus 1 (VDV-1) AY251269. Secoviridae: parsnip yellow fleck virus (PYFV) D14066, rice tungro spherical virus (RTSV) M95497, cowpea severe mosaic virus (CPSMV) M83830, satsuma dwarf virus (SDV) AB009958, apple latent spherical virus (ALSV) AB030940. Picornaviridae: poliovirus (PV) VOl149, foot-and-mouth disease virus (FMDV) X00871, encephalomyocarditis virus (EMCV) M81861, hepatitis A virus (HAV) M14707. Caliciviridae: rabbit hemorrhagic disease virus (RHDV) M67473, feline calicivirus (FCV) M86379. Marnaviridae: Heterosigma akashiwo RNA virus (HaRNAv) AY337486. Unassigned insect RNA viruses; kelp fly virus (KFV) DQ112227, Acyrthosiphon pisum virus (APV) AF0245 14.