Family: Dicistroviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are roughly spherical with a particle diameter of approximately 30 nm and no envelope (Figure 1). The virions exhibit icosahedral, pseudo T=3 symmetry and are composed of 60 protomers, each comprised of a single molecule of each of VP2, VP3 and VP1 (Figure 1). A smaller protein, VP4, is also present in the virions of some members and is located on the internal surface of the 5-fold axis below VP1.
Physicochemical and physical properties
Virions are stable in acidic conditions (to pH=3.0). Virions have a buoyant density of 1.34–1.39 g ml−1 in CsCl in the pH range of 7 to 9 and sedimentation coefficients of between 153 and 167S. However, physicochemical properties have not been fully established for all members of the family.
Nucleic acid
Virions contain a single molecule of infectious, linear, positive sense, single stranded RNA (ssRNA) of approximately 8500–10,000 nt in size with a GC content ranging from 35 to 45%. RNA constitutes about 30% of the virion weight. A small genome-linked virus protein (VPg), is covalently attached to the 5′ end of the genome. A 5′ UTR is followed by two open reading frames (ORF 1 and ORF 2) of approximately 5500 and 2500 nt, respectively. The ORFs are separated by an intergenic untranslated region (IGR) of about 170–530 nt. A UTR is also found at the 3′ end of the genome. The 3′ end of the viral RNA genome is polyadenylated. The length of the UTRs at each end of the genome is variable.
Proteins
Proteins account for 70% of the virion weight. The approximately 200 kDa nonstructural polyprotein and 100 kDa structural polyprotein are encoded by ORF 1 and ORF 2, respectively. Virions contain three major structural (capsid) viral proteins, VP1, VP2 and VP3. The size of these capsid proteins ranges from 24 to 40 kDa; an exception is Taura syndrome virus (TSV) in which VP1 is 55 kDa. A fourth smaller capsid protein (VP4) of around 4.5–9 kDa also has been reported in some species. In most species a protein precursor (VP0) is present which is cleaved to yield capsid proteins VP3 and VP4.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The RNA genome is monopartite and dicistronic with two non-overlapping ORFs that are separated and flanked by UTRs (Figure 2). The 5′-proximal and 3′-proximal ORFs encode non-structural and structural protein precursors, respectively. Components of the non-structural polyprotein include an RNA-dependent RNA polymerase (RdRp), cysteine protease and RNA helicase. The VPg attached covalently at the 5′ end of the genome plays an important role in RNA replication. The VPg sequence is repeated in most dicistrovirus genomes with the number of repeats being species-dependent. The presence of multiple VPgs is predicted to allow for multiple progeny RNA molecules to be produced per template.
Replication occurs exclusively in the cytoplasm of infected cells. Translation of the dicistronic RNA genome of dicistroviruses proceeds directly from two distinct Internal Ribosome Entry Sites (IRESs) located within the 5′-UTR and the IGR. The IGR IRES can initiate translation without codon-anticodon base pairing between the initiation AUG codon and the initiator Met-tRNA. Moreover, the IGR IRES can directly bind and manipulates the ribosome in the absence of canonical initiation factors. The activities of the IGR IRES depend on conserved RNA sequences and structures. The predicted pseudo-knot and multiple stem-loop structures in the IGR IRES are highly conserved across all members of the family and appear to be necessary for interactions with the ribosome. Conversely, the 5′-UTR IRES is not well conserved within the group and there are no clear structural homologies between the 5′-UTR IRES and IGR IRES. Translation activity of the IGR IRES is comparatively greater than that of the 5′-UTR IRES.
Antigenic properties
Many members of the family are serologically distinguishable.
Biological properties
All members of the family infect invertebrates. Most members of the family are widely distributed in nature. Aggregates and crystalline arrays of virus particles have been found in the cytoplasm of infected cells. Dicistrovirus infection is not usually associated with overt disease although infection commonly leads to reduced life expectancy. Different virus isolates vary considerably in virulence and pathogenicity; the severity of disease can range from lethal through acute to chronic and inapparent.
Species demarcation criteria in the family
The species demarcation criteria are:
- Natural host range: species can be differentiated on the basis of their natural host range and their relative ability to replicate in a range of cultured insect cells.
- Serology: species are serologically distinct.
- Sequence identity between the capsid proteins of isolates and strains of a species is above 90%.
Genus Cripavirus
Type species Cricket paralysis virus
Distinguishing features
The typical IGR-IRES elements of cripaviruses possess a conserved bulge sequence (UGAUCU and UGC) in the 5′ region (Figure 3). There is no additional stem loop presented in the 3′ region of the IGR IRES, as seen in aparaviruses.
Biological properties
Cricket paralysis virus has the widest host range among members of the genus and can infect insect species classified in the orders Diptera, Lepidoptera, Orthoptera, Hemiptera and Hymenoptera, as well as a diverse range of cultured insect cells. Other viruses have host ranges restricted to families of only one insect order. Drosophila C virus has been isolated from dipteran species. Black queen cell virus has been isolated from hymenopteran species. Aphid lethal paralysis virus, Himetobi P virus, Homalodisca coagulata virus-1, Plautia stali intestine virus, Rhopalosiphum padi virus (RhPV) and Triatoma virus (TrV) have been isolated from different families within the Hemiptera. TrV infects Triatoma infestans, a major vector of Chagas’ disease of humans. RhPV can be transmitted vertically between aphids and horizontally via the plant, using the plant as a passive reservoir. RhPV does not replicate in the plant.
List of species in the genus Cripavirus
| Aphid lethal paralysis virus |
|
|
| Aphid lethal paralysis virus | [AF536531] | (ALPV) |
| Black queen cell virus |
|
|
| Black queen cell virus | [AF183905] | (BQCV) |
| Cricket paralysis virus |
|
|
| Cricket paralysis virus | [AF218039] | (CrPV) |
| Drosophila C virus |
|
|
| Drosophila C virus | [AF014388] | (DCV) |
| Himetobi P virus |
|
|
| Himetobi P virus | [AB017037] | (HiPV) |
| Homalodisca coagulata virus-1 |
|
|
| Homalodisca coagulata virus-1 | [DQ288865] | (HoCV-1) |
| Plautia stali intestine virus |
|
|
| Plautia stali intestine virus | [AB006531] | (PSIV) |
| Rhopalosiphum padi virus |
|
|
| Rhopalosiphum padi virus | [AF022937] | (RhPV) |
| Triatoma virus |
|
|
| Triatoma virus | [AF178440] | (TrV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cripavirus but have not been approved as species
None reported.
Genus Aparavirus
Type species Acute bee paralysis virus
Distinguishing features
The members of the genus have distinguishing bulge sequence (UGGUUACCCAU and UAAGGCUU) in the region of the IGR IRES (Figure 3) and an additional stem loop in the 3′ region.
Biological properties
Except for TSV isolated from penaeid shrimps, all members of the genus infect hymenopteran species. Acute bee paralysis virus (ABPV), Israeli acute paralysis virus (IAPV) and Kashmir bee virus (KBV) are pathogens of honey bees. Consequently, these viruses can have significant detrimental effects on agricultural production by reducing pollination by honey bees. As a result of intensive international trade of honey bees and hive-associated products, these viruses have a global distribution. The parasitic mite Varroa destructor, is an effective vector for transmission of ABPV, IAPV and KBV to honey bees. Solenopsis invicta virus-1 was isolated from the red imported fire ant.
List of species in the genus Aparavirus
| Acute bee paralysis virus |
|
|
| Acute bee paralysis virus | [AF150629] | (ABPV) |
| Israeli acute paralysis virus |
|
|
| Israeli acute paralysis virus | [EF219380] | (IAPV) |
| Kashmir bee virus |
|
|
| Kashmir bee virus | [AY275710] | (KBV) |
| Solenopsis invicta virus-1 |
|
|
| Solenopsis invicta virus-1 | [AY634314] | (SINV-1) |
| Taura syndrome virus |
|
|
| Taura syndrome virus | [AF277675] | (TSV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Aparavirus but have not been approved as species
None reported.
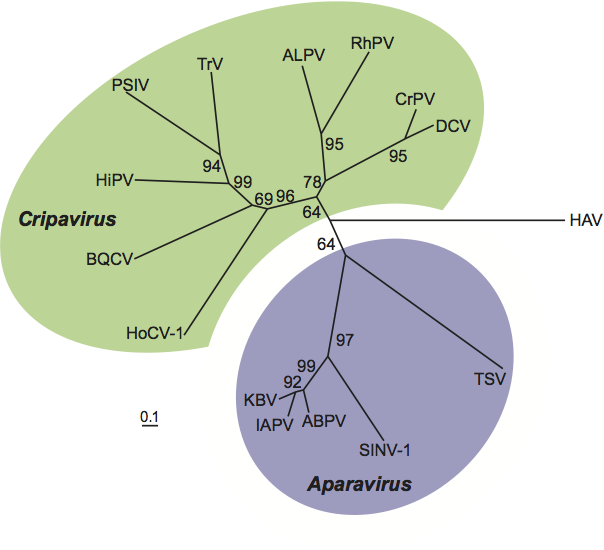
Phylogenetic relationships within the family Dicistroviridae
Phylogenetic analysis of deduced amino acid sequences of capsid protein precursors indicates that cripaviruses and aparaviruses form distinct groups in the family (Figure 4).
Similarity with other taxa
Members in the family Dicistroviridae have similarity to viruses in the Iflaviridae, Picornaviridae, Marnaviridae and Secoviridae. The genomes of viruses of these taxa are positive sense ssRNAs with a VPg and a poly (A) tail, and are translated into autoproteolytically processed polyprotein(s). Nonstructural proteins contain sequence motifs for helicase (Hel), 3C-like cysteine proteinase (Pro) and RNA-dependent RNA polymerase (RdRp) with the characteristic gene order: (Hel)-(Pro)-(RdRp). Virions contain capsid proteins organized in a module containing three related jelly-roll domains which form non-enveloped, isometric particles of pseudo T=3 symmetry and 30 nm diameter.
Derivation of names
Dicistro: from the characteristic di-cistronic arrangement of the genome.
Cripa: from the name of the type member of the genus, Cricket paralysis virus
Apara: from the type member of the genus Acute bee paralysis virus.
Further reading
Bonning and Miller, 2010 B.C. Bonning, W.A. Miller, Dicistroviruses. Annu. Rev. Entomol. 55 (2010) 129–150.
Gall et al., 2008 O.L. Gall, P. Christian, C.M. Fauquet, A.M.Q. King, N.J. Knowles, N. Nakashima, G. Stanway, A.E. Gorbalenya, Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T=3 virion architecture. Arch. Virol. 153 (2008) 715–727.
Contributed by
Chen, Y.P., Nakashima, N., Christian, P.D., Bakonyi, T., Bonning, B.C., Valles, S.M. and Lightner, D.
Figures
Figure 1 (Left) Rendering of a particle of an isolate of cricket paralysis virus (CrPV) at 2.4 resolution (Courtesy of Reddy et al.). (Center) Diagram showing the packing of surface proteins of cricket paralysis virus (CrPV). (Right) Negative contrast electron micrograph of isometric particles of CrPV. The bar represents 100 nm
(Courtesy of C. Reinganum).
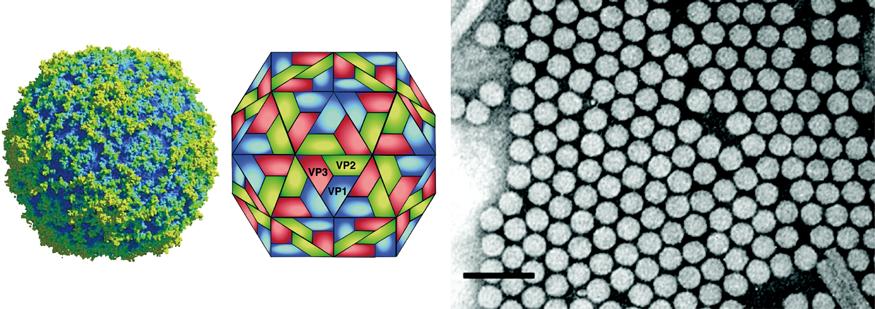
Figure 2 Genome structure of cricket paralysis virus (CrPV). The RNA genome contains two non-overlapping ORFs separated by an intergenic region (IGR). The 5 proximal ORF encodes the nonstructural proteins: RNA helicase (Hel), cysteine protease (Pro) and RNA-dependent RNA polymerase (RdRp). The structural proteins are encoded by the 3 proximal ORF and are expressed as a polyprotein that is subsequently processed to yield three major structural proteins (VP1, VP2 and VP3). VP4 is presumed to be an N-terminal extension of VP3 which is cleaved from the precursor; VP4 is considered a minor structural component of the virion. Distinct internal ribosome entry sites (IRES) are located in the 5 UTR and IGR. The genome has a small peptide covalently linked to the 5 end (genome-linked virus protein, VPg) and a 3 polyadenylated terminus.
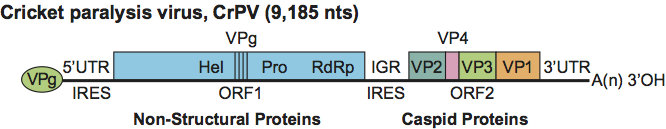
Figure 3 Comparison of IGR-IRES structural elements between cripaviruses and aparaviruses. The cripaviruses possess a conserved bulge sequence (UGAUCU and UGC) in the IGR IRES while the aparaviruses possess a bulge sequence (UGGUUACCCAU and UAAGGCUU). Additional stem loop structures and nucleotide sequences in the bulge regions are present in aparaviruses (indicated in orange).
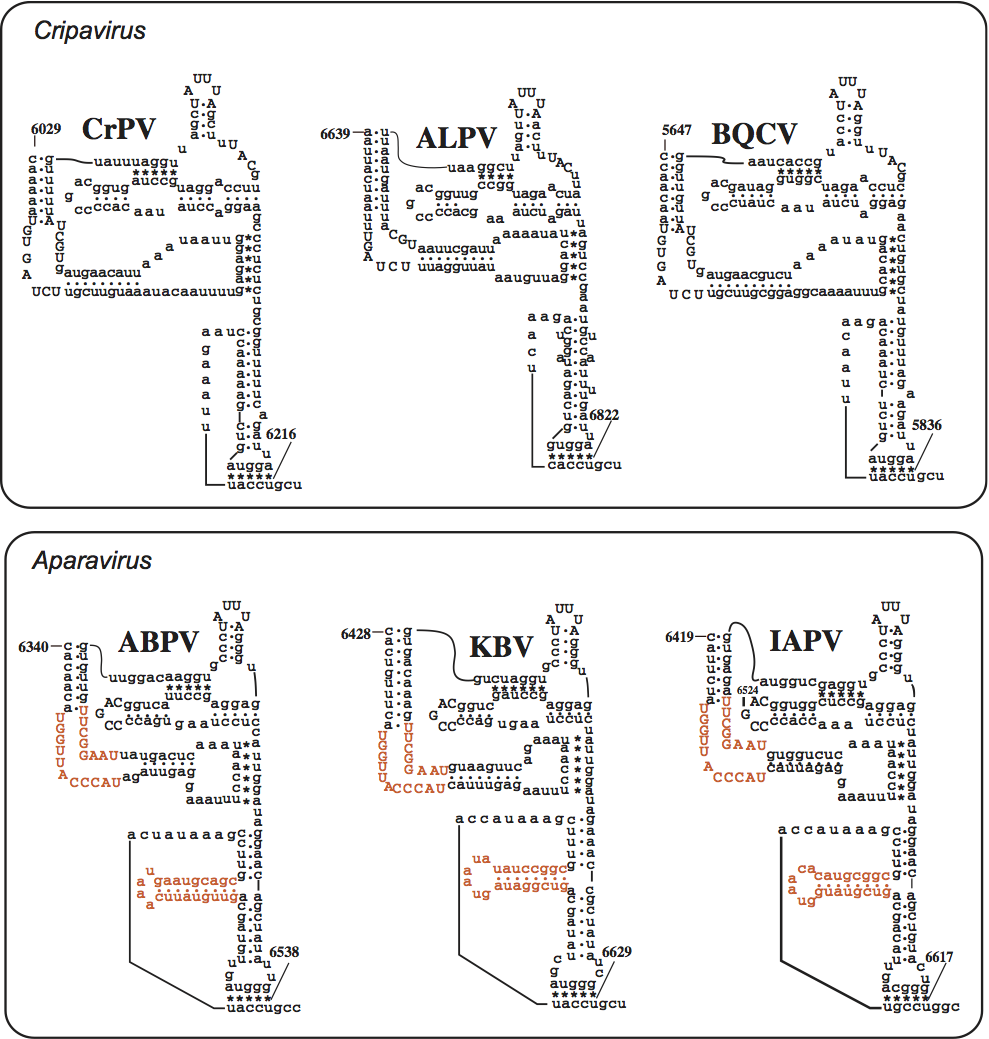
Figure 4 Neighbor-joining tree constructed from an alignment of the deduced amino acid sequence of structural proteins of dicistroviruses. Deduced amino acid sequence for capsid protein precursor of Hepatitis A virus (HAV) was used as an outgroup. Cripaviruses: ALPV, aphid lethal paralysis virus, AF536531; BQCV, black queen-cell virus, AF183905; CrPV, cricket paralysis virus, AF218039; DCV, Drosophila C virus, AF014388; HiPV, Himetobi P virus, AB017037; HoCV-1, Homalodisca coagulata virus-1, DQ288865; PSIV, Plautia stali intestine virus, AB006531; RhPV, Rhopalosiphum padi virus, AF022937; TrV, Triatoma virus, AF178440. Aparaviruses; ABPV, acute bee paralysis virus, AF150629; IAPV, Israeli acute paralysis virus, EF219380; KBV, Kashmir bee virus, AY275710; SINV-1, Solenopsis invicta virus-1, AY634314; TSV, Taura syndrome virus, AF277675.