Family: Roniviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Okavirus
Type species Gill-associated virus
Virion properties
Morphology
Virions are enveloped, bacilliform in shape with rounded ends, and 40–60 nm ×150–200 nm in dimensions (Figure 1). The envelope is studded with prominent peplomers projecting approximately 11 nm from the surface. Nucleocapsids are 20–30 nm in diameter and have a helical symmetry with coil periodicity of 5–7 nm. Long filamentous nucleocapsid precursors (approximately 15 nm in diameter and 80–450 nm in length) occur in the cytoplasm of infected cells and acquire envelopes by budding at endoplasmic reticulum membranes. Newly budded mature enveloped virions often appear as multimers butted end-to-end and paracrystalline arrays of virions can occur within vesicles.
Physicochemical and physical properties
Virion buoyant density in sucrose is 1.18–1.20 g cm-3. Yellow head virus (YHV) is inactivated by heating at 60 °C for 15–30 minutes but has been reported to survive in 25 °C–28 °C seawater for at least four days. Virions are sensitive to calcium hypochlorite and sodium dodecyl-sulfate but sensitivity to other treatments is not known.
Nucleic acid
Virions contain a single linear segment of positive sense ssRNA varying in length from 26,235 nt for gill-associated virus (GAV) to 26,662 nt for YHV. The genome contains a 5′-terminal 7-methylguanosine cap and a 3′-polyadenylated tail.
Proteins
YHV virions contain a 20–22 kDa nucleoprotein (p20) that forms the nucleocapsid and two envelope glycoproteins of 110–135 kDa (gp116) and 63–67 kDa (gp64) that form the prominent peplomers on the virion surface. The mature gp116 and gp64 glycoproteins originate by post-translational cleavage of a precursor polyprotein (pp3) encoded by the ORF3 gene. Cleavage occurs at the C-terminal side of the third and fifth of six hydrophobic transmembrane (TM) domains, which display characteristics of signal peptidase type 1-like sites. The glycoproteins gp116 and gp64 are anchored to the virion envelope by either two (TM4 and TM5) or one (TM6) transmembrane domain, respectively. There is no evidence that gp116 and gp64 are linked by intermolecular disulfide bonds.
Lipids
Virions possess a lipid envelope derived from endoplasmic membranes of host cells.
Carbohydrates
N-linked glycosylation sites are present in the amino acid sequences of gp116 (7–8 sites) and gp64 (four sites), and in virions, both are glycosylated extensively. In YHV, all but one of the seven sites in gp116 has been shown to be occupied, and three of four sites in gp64 are occupied. Carbohydrates attached to gp64 and gp116 comprise primarily mannose-type glycans, and those on gp116 also comprise N-acetyl-galactosamine and N-acetyl-glucosamine in lesser abundance. Few if any of the multiple O-linked glycosylation sites in each glycoprotein appear to be utilized.
Genome organization and replication
The positive sense ssRNA genome contains, in order from the 5′ terminus, a large replicase gene (ORF1a/ORF1b) followed by genes encoding a p20 nucleoprotein (ORF2), a pp3 precursor polyprotein (ORF3), from which the gp116 and gp64 envelope glycoproteins are derived, and a small putative protein (ORF4) of unknown function that is not translated in abundance (Figure 2). The overlap between ORF1a and ORF1b contains a slippery sequence (AAAUUUU) followed by a complex RNA pseudoknot structure causing a −1 ribosomal frameshift, with an estimated efficiency of about 23%, that allows read-through translation of the pp1a polyprotein encoded by ORF1a to generate a pp1ab polyprotein encoded by ORF1a and ORF1b. The ORF1a coding sequence contains four regions containing multiple hydrophobic domains (HD1-HD4) a chymotrypsin-like cysteine (3C-like) proteinase (3CLpro or “main” protease, Mpro) flanked by HD3 and HD4 that is involved in autolytic processing of the pp1ab polyprotein. The 3CLpro of GAV has a tentative consensus VxHE(L,V) cleavage site specificity that distinguishes it from the corresponding Mpro enzymes in vertebrate nidoviruses, and taken together with sequence differences, it appears to combine the Cys-His catalytic dyad of the coronavirus Mpro with a potyvirus-like substrate binding pocket. ORF1a also contains two putative papain-like proteinase domains (PLP1 and PLPx) that share remote structural similarity, but very little direct sequence identity, with the PLP domains of coronaviruses. Like the corresponding papain-like proteases (PLpro) of toroviruses, both PLP1 and PLPx lack a Zn2+-binding finger. In vertebrate nidoviruses, the 3CLpro and PLpros process the pp1a/pp1ab polyproteins into functional nonstructural proteins (nsp’s) numbered from the N-terminus as nsp1 to nsp12 in arteriviruses and nsp1 to nsp16 in coronaviruses. Whilst the 3CLpro of GAV has been shown to cleave at an immediate upstream site and at a site toward the pp1ab C-terminus, the number and composition of nsps generated from pp1ab by the 3CLpro in combination with the putative PLpros is not yet known. ORF1b also contains multiple sequence motifs with homologues in vertebrate nidoviruses. These include a large RdRp polymerase domain with an “SDD” active site motif and putative multinuclear Zn-finger-like domains preceding a 5′-to-3′ helicase domain (Zn-HEL), as well as homologs of the 3′-to-5′exoribonuclease (ExoN), nidoviral endoribonuclease specific for uridylate (NendoU) and ribose-2′-O-methyltransferase (O-MT) domains that exist in the C-terminal region of pp1ab replicase polyproteins of the large vertebrate nidoviruses.
Compared to the genomes of vertebrate nidoviruses, the okavirus genome is unique in that the nucleoprotein gene (ORF2) is located upstream of the glycoprotein gene (ORF3), there is no discrete gene encoding a structural membrane (M) protein, and the virion envelope glycoproteins gp116 and gp64 are generated by proteolysis of a pp3 precursor polyprotein encoded by the ORF3 gene. The approximately 25.2 kDa polypeptide cleaved from the pp3 N-terminus contains three predicted TM domains and N-linked glycosylation sites in its predicted ectodomain. Whilst this triple-membrane-spanning glycoprotein is similar in size to the M protein of some vertebrate nidoviruses, it is not abundantly present in infected cells and evidence of its association with virions in very low amounts is tenuous. Compared to the 249-nt ORF4 in GAV, ORF4 in other okavirus genotypes studied so far is truncated substantially due to deletions or sequence changes interrupting the reading frame. There is evidence of ORF4 being expressed at low levels in tissues of GAV-infected shrimp, but whether the ORF4 protein has a specific function remains unknown.
In okavirus-infected cells, two subgenome-sized (sg) mRNAs are produced, sg mRNA2 and sg mRNA3, from which ORF2 and ORF3 are translated, respectively. The genome length RNA (g RNA1) and the two sg mRNAs are 3′-coterminal and each possesses a 5′-7-methylguanosine cap structure and a 3′-poly(A) tail (Figure 2). It is likely that okavirus sg mRNAs are transcribed from complementary sg(-) strand templates as is the case in other nidoviruses. In support of this, double strand (ds) RNAs equivalent in size to the genomic and two sg mRNAs have been detected in shrimp cells infected with GAV, and likely represent replicative-intermediate RNAs. Unlike the sg mRNAs of corona- and arteriviruses, however, the okavirus sg mRNAs do not possess a common leader sequence identical to the genome 5′-terminal sequence. In contrast, g RNA1, sg mRNA2 and sg mRNA3 each initiate with a 5′AC-dinucleotide. For the sg mRNAs, this 5′-terminal AC-dinucleotide maps precisely to a central position of a sequence (GGUCAAUUACAACCUA) that is conserved in the intergenic regions (IGRs) preceding the ORF2 and ORF3 genes. Presumably, this conserved element serves a dual function, in the genome as a terminator of minus-strand RNA synthesis and in the resulting minus-strand sg RNA templates, as a transcriptional promoter for sg mRNA synthesis. Thus, okaviruses would differ from corona- and arteriviruses by not using a discontinuous transcription process to produce their mRNAs, but rather a “continuous” transcription strategy similar to that utilized by members of the subfamily Torovinae (see family Coronaviridae chapter). Interestingly, a putative terminator/promoter element is also present in the IGR preceding ORF4. However, in all genotypes studied so far, the putative transcription initiation site nucleotide is either a G or U rather than an A, which based on the absence of a sg mRNA initiating from this element, appears to destroy its functioning.
Antigenic properties
Not known.
Biological properties
Okaviruses have been detected only in crustaceans. The giant tiger shrimp (Penaeus monodon) appears to be the primary natural host of GAV and YHV but other penaeid and crustacean species are susceptible to natural and experimental infection. The geographic range of okaviruses mirrors that of its primary penaeid host and extends across the Indo-Pacific from Eastern Africa, Southern and Southeast Asia, Australia to islands in the South Pacific. Infections have also been detected recently in blue shrimp (P. stylirostris) from the Gulf of California. Infections may be chronic or acute and transmitted horizontally or vertically. The prevalence of subclinical infection in P. monodon can be high in some regions, with pathology limited to the lymphoid organ. In acute infections associated with disease and mortalities, virus invades most tissues of ectodermal and mesodermal origin. Only genotypes 1 (YHV) and 2 (GAV) have been associated with disease. High virulence is retained by a variant of genotype 1 (genotype 1b) with a 54 amino acid (aa) deletion at the gp116 N-terminus, although its LD50 is somewhat lower than the highly-virulent genotype 1a virus.
Species demarcation criteria in the genus
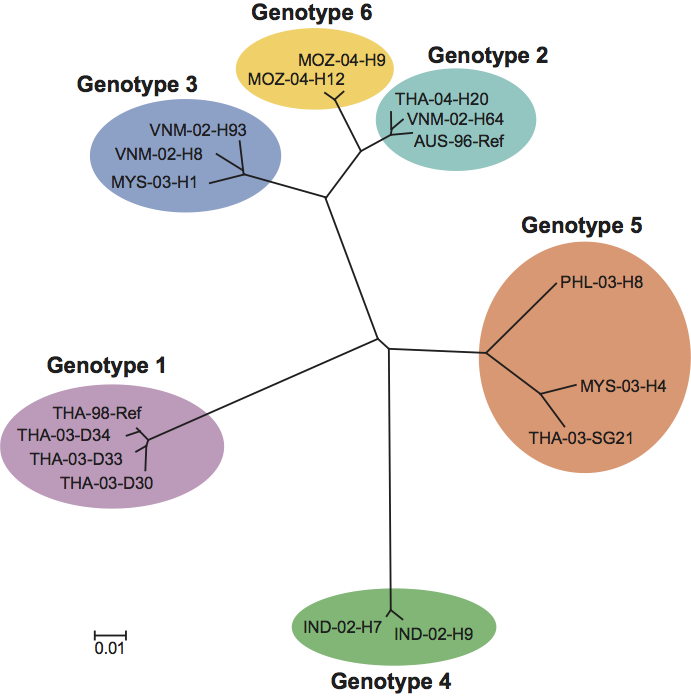
In the genus Okavirus, differences in genome organization and biological properties of the six assigned genotypes detected in P. monodon shrimp appear insufficient to warrant their demarcation into separate species. Evidence of genetic recombination between genotypes also indicates that all should be classified as a single species.
List of species in the genus Okavirus
| Gill-associated virus |
|
|
| Gill-associated virus - Australia | [AF227196] | (GAV-AUS) |
| Yellow head virus | [FJ848673] | (YHV) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Okavirus but have not been approved as species
None reported.
Phylogenetic relationships within the family
Phylogenetic analyses of a partial ORF1b gene sequence has clustered okaviruses into six distinct genotypes numbered 1 (YHV), 2 (GAV), 3, 4, 5 and 6 (Figure 3). Genotype 1, 2, 3 and 5 viruses have been detected in P. monodon from various locations in Southeast Asia, but genotypes 2, 4 and 6 are the sole genotypes thus far detected in P. monodon from Australia, India and eastern Africa, respectively. There is evidence of naturally occurring genetic recombination involving genotypes 1 (YHV), 2 (GAV), 3 and 5.
Similarity with other taxa
Similarities in the genome size and organization, sequences of core replicase proteins and the sgmRNA transcription strategy used by roniviruses clearly link them to other members of the order Nidovirales. The RNA pseudoknot component of the −1 ribosomal frameshift in the ORF1a/1b gene overlap resembles the gag/pol pseudoknots of some retroviruses. The structure and substrate specificity of the pp1ab chymotrypsin-like cysteine proteinase (3CLpro) bridges phylogenetic distinctions between the corresponding proteinases of coronaviruses and plant potyviruses.
Derivation of names
Roni: from rod-shaped nidovirus referring to the virion morphology of viruses in the family.
Oka: refers to the lymphoid or so named “oka” organ, an anatomical structure common to penaeid shrimp, and which is a site of virus replication and pathology in both subclinical and acute infections.
Further reading
Cowley et al., 2000 J.A. Cowley, C.M. Dimmock, K.M. Spann, P.J. Walker, Gill-associated virus of Penaeus monodon shrimp: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81 (2000) 1473–1484.
Cowley et al., 2001 J.A. Cowley, C.M. Dimmock, P.J. Walker, Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic RNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83 (2001) 927–935.
Cowley and Walker, 2002 J.A. Cowley, P.J. Walker, The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 147 (2002) 1977–1987.
Cowley and Walker, 2008 J.A. Cowley, P.J. Walker, S. Perlman, T. Gallagher, E.J. Snijder, Molecular biology and pathogenesis of ronivirusesNidoviruses. In: S. Perlman, T. Gallagher, E.J. Snijder, Nidoviruses. ASM Press, Washington, DC2008361–377.
Jitrapakdee et al., 2003 S. Jitrapakdee, S. Unajak, N. Sittidilokratna, R.A.J. Hodgson, J.A. Cowley, P.J. Walker, S. Panyim, V. Boonsaeng, Identification and analysis of gp116 and gp64 structural glycoproteins of yellow head nidovirus of Penaeus monodon shrimp. J. Gen. Virol. 84 (2003) 863–873.
Sittidilokratna et al., 2008 N. Sittidilokratna, S. Dangtip, J.A. Cowley, P.J. Walker, RNA transcription analysis and completion of the genome sequence of yellow head nidovirus. Virus Res. 136 (2008) 157–165.
Sittidilokratna et al., 2002 N. Sittidilokratna, R.A.J. Hodgson, J.A. Cowley, S. Jitrapakdee, V. Boonsaeng, S. Panyim, P.J. Walker, Complete ORF1b-gene sequence indicates yellow head virus is an invertebrate nidovirus. Dis. Aquat. Organ. 50 (2002) 87–93.
Spann et al., 1997 K.M. Spann, J.A. Cowley, P.J. Walker, R.J.G. Lester, Gill-associated virus (GAV), a yellow head-like virus from Penaeus monodon cultured in Australia. Dis. Aquat. Organ. 31 (1997) 169–179.
Wijegoonawardane et al., 2008 P.K.M. Wijegoonawardane, J.A. Cowley, T. Phan, R.A.J. Hodgson, L. Nielsen, W. Kiatpathomchai, P.J. Walker, Genetic diversity in the yellow head nidovirus complex. Virology. 380 (2008) 213–225.
Wijegoonawardane et al., 2009 P.K.M. Wijegoonawardane, N. Sittidilokratna, N. Phetchampai, J.A. Cowley, N. Gudkovs, P.J. Walker, Homologous genetic recombination in the yellow head complex of nidoviruses infecting Penaeus monodon shrimp. Virology. 390 (2009) 79–88.
Ziebuhr et al., 2003 J. Ziebuhr, S. Bayer, J.A. Cowley, A.E. Gorbalenya, The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 77 (2003) 1415–1426.
Contributed by
Cowley, J.A., Walker, P.J., Flegel, T.W., Lightner, D.V., Bonami, J.R., Snijder, E.J. and de Groot, R.J.
Figures
Figure 1 (A) Transmission electron micrograph of negative-stained virion of gill-associated virus (GAV). (B) Transmission electron micrograph of partially disrupted yellow head virus (YHV) virion displaying the internal nucleocapsid and a ring-like structure which appears to be a disrupted virion in cross-section. (C) Schematic illustration of an okavirus virion. (D) Transmission electron micrograph of unenveloped cytoplasmic nucleocapsids in a thin section of GAV-infected shrimp cells. The bars represent 100 nm.
(Electron micrographs provided by K.M. Spann, P. Loh, J.A. Cowley and R.J. McCulloch and reproduced with permission.)
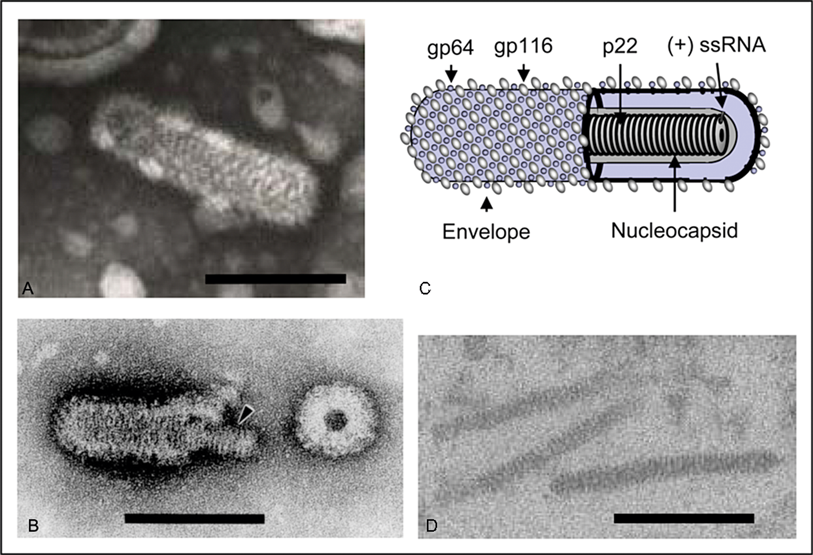
Figure 2 Schematic representation of the 26,662 nt polyadenylated (+) ssRNA genome of YHV (g RNA1) and the two 3-coterminal sub-genomic RNAs sg mRNA2 and sg mRNA3. Functional domains in ORF1a: hydrophobic regions (HD1-HD4), 3C-like protease (3CLP), papain-like protease (PLP1) and a domain with homology to PLP1 but lacking the canonical + fold of papain-like proteases (PLPx). Functional domains in ORF1b: RNA polymerase (RdRp), cysteine- and histidine-rich zinc finger and helicase (Zn-Hel), exoribonuclease (ExoN), uridylate-specific endoribonuclease (N) and ribose-O-methyl transferase (MT). ORF2 encodes the nucleoprotein (p20). ORF3 encodes a precursor polyprotein (pp3) that undergoes post-translation processing to generate envelope glycoproteins (gp116 and gp64) and an N-terminal triple-membrane-spanning fragment of unknown function (p25). The ribosomal frameshift site (RFS) allows read-through translation of polyprotein pp1ab from ORF1a and ORF1b. Known () and likely () sites of proteolytic cleavage of expressed polyproteins pp1a and pp1ab are indicated together with the two signal peptidase type 1 cleavage sites in pp3 ().
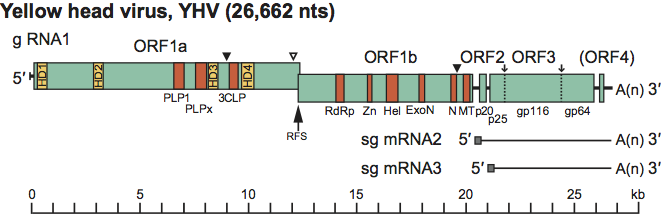
Figure 3 Unrooted phylogenetic tree of representatives of each of the six okavirus genotypes based on a 781 nucleotide region of the ORF1b gene [see Wijegoonawardane et al. (2008) for strain details and expanded phylogenetic analyses]. The sequences were aligned using Clustal W, phylogenetic inference was determined by the neighbor-joining method and the tree was constructed using Treeview software. Bootstrap values obtained for a 1000 replicate analyses were 1000 at each genotype branching node. Branch lengths are proportional to phylogenetic distance and the bar represents a sequence divergence of 1.0%.