Family: Arteriviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Arterivirus
Type species Equine arteritis virus
Virion properties
Morphology
Arteriviruses are pleomorphic but roughly spherical particles. By cryo-electron microscopy porcine reproductive and respiratory syndrome virus (PRRSV) particle diameters were found to range from 50 to 74 nm, with a median value of 54 nm and only few particles larger than 60 nm. Using the same approach the average diameter of the isometric nucleocapsid, which is probably not icosahedrally ordered, was found to be 39 nm. The nucleocapsid is surrounded by a lipid envelope with small surface projections that cover the entire virion surface (Figure 1).
Physicochemical and physical properties
The buoyant density of arterivirus particles has been estimated to be 1.13 to 1.17 g cm−3 in sucrose. Reported sedimentation coefficients for arteriviruses range from 200S to 300S. Virions are stable when stored at −70 °C. The half-life of arteriviruses progressively decreases with increasing temperature. Virions are stable between pH 6.0 and 7.5, but are inactivated at high or low pH. Arteriviruses are also inactivated by lipid solvents, such as ether, butanol and chloroform and are extremely sensitive to detergent treatment. A brief incubation with a nonionic detergent such as 0.01% NP40 or Triton X-100 efficiently disrupts the viral envelope.
Nucleic acid
Virions contain a single molecule of linear, positive sense, single stranded RNA that ranges in length from 12.7 to 15.7 kb (Figure 2). The naked RNA, when transfected into permissive cells, is itself infectious. The genomic RNA contains a 5′ type I cap structure (simian hemorrhagic fever virus, SHFV) and a 3′-terminal poly(A) tract. Full-length sequences are available in the GenBank database for representatives of all currently known arterivirus species.
Proteins
Seven structural proteins have been identified in equine arteritis virus (EAV) and PRRSV virions (Table 1). In addition to the nucleocapsid protein (N), there are two major (GP5 and M) and four minor (E, GP2, GP3, GP4) envelope proteins (Figures 1–3). By reverse genetics (EAV and PRRSV), each of these proteins was shown to be required for the production of infectious progeny. The major glycoprotein, GP5, spans the membrane three times and forms a disulfide-linked heterodimer with triple membrane spanning M protein; the heterodimer between conserved cysteine residues is essential for virus infectivity (Figure 3A). The GP5 proteins of EAV and SHFV are predicted to possess 98 residues on the outside of the virion, while both genotypes of PRRSV and lactate dehydrogenase-elevating virus (LDV) are predicted to have ectodomains of approximately 30 amino acids. The predicted orientation of the minor envelope proteins in the viral membrane is also shown in Figure 3B. The E protein of EAV and PRRSV is fatty-acid acylated, and the myristoylation of E has been shown to be non-essential for virus infectivity in both viruses. The E protein appears to be an ion-channel protein that may function in the uncoating process during virus entry and penetration. GP2, GP3 and GP4 form heterotrimers on the surface of the virus particle (EAV and PRRSV). The proteins encoded by ORFs 2a/b, 3 and 4 have not been confirmed as structural components of LDV, nor has the trimerization of these proteins been assessed. A soluble, non-virion associated form of the ORF3 glycoprotein is also released from infected cells (LDV and PRRSV Type 2). In addition, the N proteins of EAV and PRRSV Type 2 have been shown to dimerize. Crystal structures of the putative dimerization domain of arterivirus N proteins (PRRSV Type 2 and EAV) have been determined and indicate that these proteins represent a new class of viral capsid-forming proteins. Virus mutants (EAV) lacking expression of E, GP2, GP3 or GP4 produce non-infectious particles. The virion proteins of SHFV include the two major envelope proteins (GP5 and M) and the N protein; the genome possesses four additional 3′ ORFs (2a′, 2b′, 3′ and 4′), encoding GP2′, E′, GP3′ and GP4′, which may be duplications of ORFs 2 a/b to 4 (Figure 2).
Table 1 Virion-associated proteins of arteriviruses
| Protein | aab | ORF | mRNA | (Putative) Function(s) |
| Ed | 67–80 | 2a/2bc | 2d | Small integral envelope protein, myristoylated, postulated ion channel protein |
| GP2a/b | 227–256 | 2b/2ac | 2d | Minor glycoprotein, part of GP2/GP3/GP4 heterotrimer |
| GP3 | 163–265 | 3 | 3 | Minor glycoprotein, part of GP2/GP3/GP4 heterotrimer |
| GP4 | 152–183 | 4 | 4 | Minor glycoprotein, part of GP2/GP3/GP4 heterotrimer |
| GP5 | 199–278 | 5 | 5 | Major glycoprotein, carries main determinants for neutralization, part of GP5/M heterodimer |
| M | 162–174 | 6 | 6 | Integral membrane protein, part of GP5/M heterodimer |
| Ne,f | 110–128 | 7 | 7 | Nucleocapsid protein, partially localizes to the nucleus of infected cells, phosphoprotein |
a Based on the genome organization of EAV, PRRSV and LDV, adapted from the Eighth ICTV Report; SHFV ORFs 2a-7 were used to predict structural protein amino acid length.
b aa=amino acids.
c In PRRSV.
d Subgenomic mRNA2 is assumed to be functionally bicistronic.
e Partial crystal structures have been determined for EAV and PRRSV N (Protein Data Bank IDs 2I9F and 1P65, respectively).
f ORF7 polymorphism in Type 1 (125, 126, 129, 131 AA) and Type 2 (124, 125 AA) PRRSV.
Lipids
The virion lipids are cell derived, with virus budding occurring on membranes of the ER and the Golgi part of the exocytic pathway.
Carbohydrates
Although the number of predicted N-linked glycosylation sites in GP2-5 of EAV, PRRSV and LDV strains differ, the large majority of these sites are highly conserved and present in almost all natural isolates of the three viruses. Specifically, it has been demonstrated that conserved N-linked glycosylation sites in GP5 are essential for virus infectivity and antigenicity (EAV and PRRSV). Recent reverse genetics studies have revealed that PRRSV and EAV mutants void of GP5 N-linked glycosylation are nonviable or readily revert to restore at least one N-linked glycosylation site. Since stable virus mutants lacking these carbohydrates cannot be generated, further investigation of the specific role of glycans is difficult. However, a viable pseudorevertant (deletion mutant) of an EAV mutant with a completely unglycosylated GP5 ectodomain was isolated. This pseudorevertant grew to a lower titer and produced smaller plaques compared to the parental virus with a single conserved GP5 glycosylation site. The sugar composition of the N-linked glycans of GP2-5 of these arteriviruses has not been investigated in detail. Preliminary studies indicate that the EAV GP5 carbohydrate is modified with N-acetyllactosamine, while PRRSV GP5 is thought to contain complex sugars other than polylactosaminoglycans.
Antigenic properties
Each of the four arteriviruses is antigenically distinct and arteriviruses show no serological cross-reactivity. Furthermore, there is significant antigenic variation among different strains of EAV, LDV and PRRSV, but the antigenic properties of SHFV have not been evaluated. The major neutralization epitopes of arteriviruses have been mapped to the GP5 protein of EAV, PRRSV (both Type 1 and 2) and LDV and SHFV 5. It has been shown that GP5 and M heterodimerization is critical for the expression of neutralization epitopes on GP5 proteins of EAV and PRRSV (Figure 3A). In contrast to Type 2 PRRSV, neutralizing antibodies against GP4 have also been shown to neutralize the Type 1 PRRSV prototype (Lelystad). In addition to GP5, the M protein is the most frequently recognized EAV structural protein by convalescent sera from non-persistently infected horses, whereas sera from carrier stallions also recognize the GP3 and N proteins. The antigenicity of and humoral antibody response to EAV nsps have yet to be determined. Antibodies to several nonstructural proteins (nsps; nsp1, nsp2 and nsp7), N and M proteins and GP5 appear within 7 to 14 days of PRRSV infection. However, the neutralizing antibody response against GP5 is delayed. A number of B cell epitopes have been identified in the nsp2 protein of PRRSV. Similarly, a number of T-cell epitopes have been identified on GP4, GP5, M and N proteins of PRRSV. Infection with PRRSV also leads to suppression of T lymphocytes, and a number of nsps have been implicated in countering host immune responses. Infection with some PRRSV strains result in a transient induction of high levels of serum interferon gamma. Mice infected with LDV develop a strong neutralizing antibody response to GP5 early in the infection that effectively neutralizes the virus. Nevertheless, virus isolated from persistently infected mice is neutralization-resistant, suggesting selection of neutralization escape variants. The activation of T lymphocytes by LDV infection is limited to the first day postinfection and is triggered by the large amounts of interferon alpha produced by the initial productively-infected macrophages. This T-lymphocyte activation is rapidly followed by a transient suppression of T cell responses (cytotoxic and helper) that is maximal at about 3 days postinfection. LDV and PRRSV (in gnotobiotic pigs) also trigger polyclonal B-cell activation.
Arterivirus entry
The arterivirus cell tropism has been attributed to the presence or absence of specific entry mediators. The entry of PRRSV into the porcine macrophage has been intensively studied but little is yet know about the entry of EAV, LDV and SHFV into their respective target cells. PRRSV virions adhere to macrophages through interactions with heparan sulphate glycosaminoglycans that line the cell surface. Subsequently, the virus binds to the macrophage-specific lectin sialoadhesin. Sialic acids on the virion surface are crucial for this interaction and the M/GP5 glycoprotein complex was identified as a ligand for this lectin receptor. Binding of PRRSV to sialoadhesin triggers uptake of the virus-receptor complex via clathrin-mediated endocytosis and subsequent release of the viral genome into the cytoplasm of the target cell initiates the translational and transcriptional processes that lead to productive infection. PRRSV genome release is dependent on acidification of the virus-containing endosome, and both cellular CD163 and proteases (cathepsin E) appear to play a crucial role in this step. GP2 and GP4 were identified as binding partners for CD163 by co-precipitation experiments, indicating a potential function for these molecules in genome release.
Genome organization and replication
For EAV, PRRSV and LDV, the genome contains nine functional ORFs, whereas the single reported SHFV sequence contains 13 ORFs, due to an apparent 4-gene duplication (Figure 2). The viral genes for EAV, PRRSV and LDV are arranged in the order 5′-replicase-E/GP2-GP3-GP4-GP5-M-N-3′, and mostly overlap. A schematic of the arterivirus replication cycle, based on studies with EAV, is shown in Figure 4. Following genome translation, dedicated nonstructural proteins (nsps) are thought to induce double-membrane vesicles (DMVs) with which the replication/transcription complexes of arteriviruses become associated in order to engage in RNA-dependent RNA synthesis (Figures 2 and 4). In addition to a full-length minus strand, infected cells also contain a nested set of subgenomic minus strand RNAs that are the complements of the subgenomic mRNAs (sgmRNAs) and are believed to function as templates for their transcription. The 5′-proximal 156–211 nt of the sgmRNAs (“leader” or 5′ UTR) are derived from the 5′ end of the genome, whereas the coding region of the sgmRNA (the mRNA “body”) is colinear with the 3′-proximal region of the genome (Figure 2). The synthesis of the subgenomic minus strands, which involves the fusion of sequences that are noncontiguous in the genome, is currently thought to occur by discontinuous extension during minus strand synthesis. This is a process in which transcription-regulating sequences (TRSs) direct attenuation of minus strand synthesis, translocate the nascent strand to the leader region in the 5′ end of the genomic template, and (guided by a base-pairing interaction) reinitiate minus strand synthesis to add the leader complement to the nascent subgenomic minus strand RNA. High frequency intraspecies RNA recombination between divergent strains of EAV, LDV and PRRSV has been conclusively shown. For PRRSV, only intragenotype (Type 1 or 2) recombination has been detected to date.
Four cell proteins (36, 55, 86 and 103 kDa) have been identified that bind to a cis-acting region required for plus-strand RNA synthesis from the minus-strand template, and two cell proteins (polypyrimidine tract-binding protein and fructose 1,6 bisphosphate aldolase) have been shown to bind to the 3′ UTR of the positive strand, required for negative strand synthesis of SHFV and LDV-C. Other cell proteins have been identified that bind to the 5′ UTRs of EAV positive and negative strand RNAs.
Nonstructural proteins
The arterivirus genome is polycistronic and contains 9 to 13 ORFs. Three-quarters of the 5’-proximal length of the genome is occupied by two large ORFs that together encode all viral enzyme functions (collectively referred to as the viral “replicase”, Figure 2) required for genome replication and subgenomic mRNA production. Replicase ORFs 1a and 1b are translated into polyproteins pp1a (187–260 kDa) and pp1ab (345–421 kDa), with the latter being a C-terminally extended version of the former. ORF1b translation depends on a -1 ribosomal frameshift just before termination of ORF1a translation. A number of conserved domains are present in the replicases of all arteriviruses (from N-terminus to C-terminus; Figure 5): a zinc finger (ZF in nsp1), a papain-like cysteine proteinase (PLP1α and/or β [and PLP1γ in SHFV) in nsp1], an unusual papain-like cysteine proteinase (PLP2 or CP in nsp2), a chymotrypsin-like serine proteinase (SP in nsp4; alternatively known as serine Main protease, Mpro), a RNA-dependent RNA polymerase (RdRp in nsp9), a zinc binding domain (Z in nsp 10), a NTPase/RNA helicase (HEL in nsp10), and a nidovirus uridylate-specific endoribonuclease (NendoU; U in nsp11). The RNA polymerase (EAV), helicase (EAV and PRRSV Type 2), and endoribonuclease activities (EAV and PRRSV Type 2) of nsp9, nsp10 and nsp11, respectively, have been corroborated with biochemical assays using purified recombinant proteins. Furthermore, EAV nsp1 has been identified as a critical regulator of the accumulation levels of genomic and subgenomic mRNAs, most likely by modulating the synthesis of their corresponding minus strand templates. ORF1a-encoded subunits with hydrophobic domains, in particular nsp2 and nsp3, have been implicated in the formation of the endoplasmic reticulum-derived DMVs with which the viral replication and transcription complexes are associated. Detailed analysis has shown that the EAV replicase polyproteins pp1a and pp1ab are cleaved into 13 mature nsps by the three viral proteinases (PLP1β, PLP2 and SP) (Table 2), whereas LDV and PRRSV produce at least one additional cleavage product due to the fact that their nsp1 equivalent contains a second internal proteinase that cleaves the nsp1 region into nsp1α and nsp1β. PLP2 (CP) has been shown to cleave nsp2 from nsp3 in EAV and PRRSV, and to assist the SP in cleavage of the nsp4/5 junction in EAV (Table 2). The EAV nsp4 SP is responsible for nine proteolytic cleavages in the C-terminal half of pp1a and the ORF1b-encoded part of pp1ab. Several cysteine/histidine (C/H) motifs have been identified, and two (putative) zinc binding domains (in nsp1 and nsp 10) have been shown to be critical for the EAV replication cycle (Table 2). By sequence comparison, similar cleavage sites and C/H motifs have been predicted in all other arteriviruses, but most are not confirmed. The PLP1β cleavage site for PRRSV has been shown to be different from what was originally proposed.
Table 2 Nonstructural proteins of EAV
| Protein | aab | Mode of expressionc | (Putative) Function(s) |
| nsp1d | 260 | TI + nsp1 PLP1β | Zinc finger, proteinase (PLP1), replicase polyprotein processing, transcription, and virion biogenesis (dispensable for genome replication) |
| nsp2 | 571 | TI + nsp1 PLP1β + nsp2 PLP2 | Proteinase (PLP2) with deubiquitinating activity (DUB), integral membrane protein, replication complex (DMV) formation |
| nsp3 | 233 | TI + nsp2 PLP2 + nsp4 SP | Integral membrane protein, replication complex (DMV) formation |
| nsp4e | 204 | TI + nsp4 SP | Main proteinase (SP) |
| nsp5 | 162 | TI + nsp4 SP | Integral membrane protein, replication complex (DMV) formation |
| nsp6 | 22 | TI + nsp4 SP | ? |
| nsp7α, β | 225 | TI + nsp4 SP | ? |
| nsp8f | 50 | TI + nsp4 SP + TT | ? |
| nsp9 | 693 | TI + RFS + nsp4 SP | RNA-dependent RNA polymerase |
| nsp10 | 467 | TI + RFS + nsp4 SP | RNA helicase/NTPase, putative zinc binding domain, role in subgenomic mRNA synthesis |
| nsp11 | 219 | TI + RFS + nsp4 SP | Nidovirus uridylate-specific endoribonuclease |
| nsp12 | 119 | TI + RFS + nsp4 SP + TT | ? |
a Based on the currently known replicase processing scheme of EAV, adapted from the Eighth ICTV Report.
b aa=amino acids.
c TI, translation initiation; RFS, ORF1a/ORF1b ribosomal frameshifting; TT, translation termination; PLP1, papain-like cysteine proteinase; PLP2, papain-like, cysteine proteinase; SP, serine proteinase.
d Nsp1 of LDV and PRRSV is cleaved internally by an additional papain-like proteinase to yield nsp1α and nsp1β. The nsp1α subunit of PRRSV contains two distinct zinc finger configurations, the second of which has been shown to be functional. A crystal structure has been determined for PRRSV nsp1α (Protein Data Bank ID 3IFU).
e Crystal structures have been determined for EAV and PRRSV nsp4 (Protein Data Bank IDs 1MBM and 3FAO, respectively).
f Due to ribosomal frameshifting, nsp8 is identical to the N-terminal 50 aa of nsp9.
Biological properties
All known arteriviruses infect a single type of vertebrate host in the domain Eucarya and are not transmitted by a vector. Macrophages are the primary host cell. Arteriviruses are cytocidal and usually cause both acute and chronic, persistent disease in their hosts. EAV is the agent of equine viral arteritis (inflammation of small arteries) and infection can lead to extremely variable clinical signs. Viral arteritis has not been a reported characteristic of infection with LDV and PRRSV. LDV causes a lifelong persistent infection of mice, but CNS disease resulting in paralysis is seen in Fv-1n/n mice. Infection of swine with PRRSV results in reproductive failure in sows, respiratory illness in growing swine, and is usually asymptomatic in boars. For PRRSV, extraordinary strain diversity has been detected and is due in part to high levels of viral recombination (Type 1 and 2) that leads to variation in clinical symptoms from mild to severe (especially in Type 2). SHFV causes acute or persistent infections in African monkeys, such as patas (Erythrocebus patas), with no overt disease symptoms, but induces a fatal hemorrhagic fever in monkeys of the genus Macaca.
Species demarcation criteria in the genus
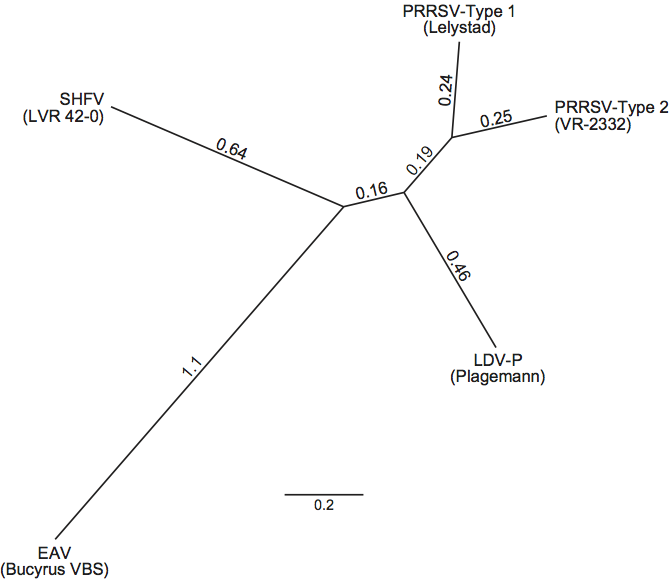
Each of the four currently recognized species in the genus Arterivirus constitutes a major phylogenetic branch within the genus (Figure 6). For each species except SHFV, multiple full-length sequences were determined and indicate that the species consist of clusters of sometimes widely divergent strains. LDV and PRRSV are most closely related to each other. There are two subspecific types of PRRSV, European (Type 1) and North American (Type 2). Members of the four virus species are antigenically distinct. The viruses of each species have a restricted host range and each species infects different hosts. The size variation of the N-terminal half of ORF1 suggests that this arterivirus region may encode species-specific functions.
List of species in the genus Arterivirus
| Equine arteritis virus |
|
|
| Equine arteritis virus - virulent Bucyrus strain (VBS) | [DQ846750] | (EAV-VBS) |
| Equine arteritis virus - cell-culture adapted Bucyrus | [X53459=NC_002532] | (EAV) |
| Lactate dehydrogenase-elevating virus |
|
|
| Lactate dehydrogenase-elevating virus-Plagemann | [U15146=NC_001639] | (LDV-P) |
| Lactate dehydrogenase-elevating virus-C | [L13298] | (LDV-C) |
| Porcine respiratory and reproductive syndrome virus |
|
|
| Porcine respiratory and reproductive syndrome virus-Type 1 | [M96262] | (PRRSV-1) |
| Porcine respiratory and reproductive syndrome virus-Type 2 | [U87392] | (PRRSV-2) |
| Simian hemorrhagic fever virus |
|
|
| Simian hemorrhagic fever virus | [AF180391=NC_003092] | (SHFV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Arterivirus but have not been approved as species
None reported.
List of unassigned species in the family Arteriviridae
None reported.
Phylogenetic relationships within the family
Phylogenetic analyses show that the family includes five major lineages, indicating that lineages represented by Type 1 and Type 2 PRRSV may be recognized as separate species in a future revision of arterivirus taxonomy (Figure 6).
Similarity with other taxa
The family Arteriviridae together with the family Coronaviridae and the family Roniviridae form the order Nidovirales. These viruses have important common features at the level of genome organization, genome expression strategy, phylogeny and internal organization of their large replicase gene. Despite these overall similarities, arterivirus genomes are substantially smaller, and the size, structure and composition of their virions do not resemble those of other members of the order Nidovirales. Consequently, arteriviruses may be called small-genome nidoviruses, while the others are recognized as large-genome nidoviruses. Various non-structural proteins contain arterivirus- and/or nidovirus-specific domains or signatures, including an SDD signature (instead of the canonical GDD) in the RdRp (nsp9), a complex N-terminal (putative) zinc binding domain in the helicase (nsp10), and a putative endonuclease domain (NendoU) in nsp11. Non-nidovirus homologs of several (putative) enzymes encoded by viruses of the family Arteriviridae have been found in other RNA viruses. The main proteinase and RdRp cluster together with homologs of viruses in the “picornavirus-like” and “sobemovirus-like” supergroups. The helicase has counterparts in viruses of the “alphavirus-like” supergroup. Also, some parallels are evident between the genome organization and expression strategy of members of the family Arteriviridae (and other members of the order Nidovirales) and those of the plant virus family Closteroviridae.
Derivation of name
Arteri: derived from the disease caused by the type member, equine arteritis virus.
Further reading
Balasuriya and MacLachlan, 2004 U.B. Balasuriya, N.J. MacLachlan, The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102 (2004) 107–129.
Chen et al., 2010 Z. Chen, S. Lawson, Z. Sun, X. Zhou, X. Guan, J. Christopher-Hennings, E.A. Nelson, Y. Fang, Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 398 (2010) 87–97.
Gorbalenya et al., 2006 A.E. Gorbalenya, L. Enjuanes, J. Ziebuhr, E.J. Snijder, Nidovirales: Evolving the largest RNA virus genome. Virus Res. 117 (2006) 17–37.
Kimman et al., 2009 T.G. Kimman, L.A. Cornelissen, R.J. Moormann, J.M. Rebel, N. Stockhofe-Zurwieden, Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine. 27 (2009) 3704–3718.
Pasternak et al., 2006 A.O. Pasternak, W.J. Spaan, E.J. Snijder, Nidovirus transcription: how to make sense…?. J. Gen. Virol. 87 (2006) 1403–1421.
Perlman et al., 2008 S. Perlman, T. Gallagher, E.J. Snijder, Nidoviruses. ASM Press, Washington, DC2008.
Plagemann and Moennig, 1992 P.G. Plagemann, V. Moennig, Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv. Virus Res. 41 (1992) 99–192.
Plagemann et al., 1995 P.G.W. Plagemann, R.R.R. Rowland, C. Even, K.S. Faaberg, Lactate dehydrogenase- elevating virus – an ideal persistent virus?. Springer Semin. Immunopathol. 17 (1995) 167–186.
Snijder and Meulenberg, 1998 E.J. Snijder, J.J. Meulenberg, The molecular biology of arteriviruses. J. Gen. Virol. 79 (1998) 961–979.
Spilman et al., 2009 M.S. Spilman, C. Welbon, E. Nelson, T. Dokland, Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J. Gen. Virol. 90 (2009) 527–535.
Van Breedam et al., 2010 W. Van Breedam, P.L. Delputte, H. Van Gorp, G. Misinzo, N. Vanderheijden, X. Duan, H.J. Nauwynck, Porcine reproductive and respiratory syndrome virus entry in the porcine macrophage. J. Gen. Virol. 91 (2010) 1659–1667.
Contributed by
Faaberg, K.S., Balasuriya, U.B., Brinton, M.A., Gorbalenya, A.E., Leung, F.C-C., Nauwynck, H., Snijder, E.J., Stadejek, T., Yang, H. and Yoo, D.
Figures
Figure 1 Structure of arterivirus virions. (A) Cryo-EM of PRRSV particles (strain SD-23983) in vitreous ice. The bar represents 100 nm. The white arrow points to a particle with a rectangular core. Black arrows indicate protruding features thought to correspond to complexes of the minor envelope proteins. Inset, magnified (2) view of a single, typical PRRSV particle with dimensions indicated. A presumed envelope spike complex is indicated, as is the striated appearance most likely corresponding to transmembrane domains. The dark area on the left is part of the carbon support film (from Spilman et al. (2009). J. Gen. Virol., 90, 527-535; with permission from the Journal of General Virology. (B) Schematic representation of the arterivirus particle. N-glycosylation sites are indicated by stars. (C) Structure of the core. (1) Cutaway view of one PRRSV virion. The envelope, shown in mesh representation, was peeled away to reveal the internal core. The core is shown as an isosurface, coloured by the radius from the centre of the particle (from red to blue). (2) The core has been cut open to show the internal structure and the characteristic central density (red-orange). (3) A 63 nm thick slab through the centre of one particle tomogram, with several copies of the crystal structure of the dimer of the C-terminal domain of N rendered at a comparable resolution to the tomogram and superimposed on the oblong densities in the core
(from Spilman et al. (2009). J. Gen. Virol., 90, 527-535; with permission from the Journal of General Virology).
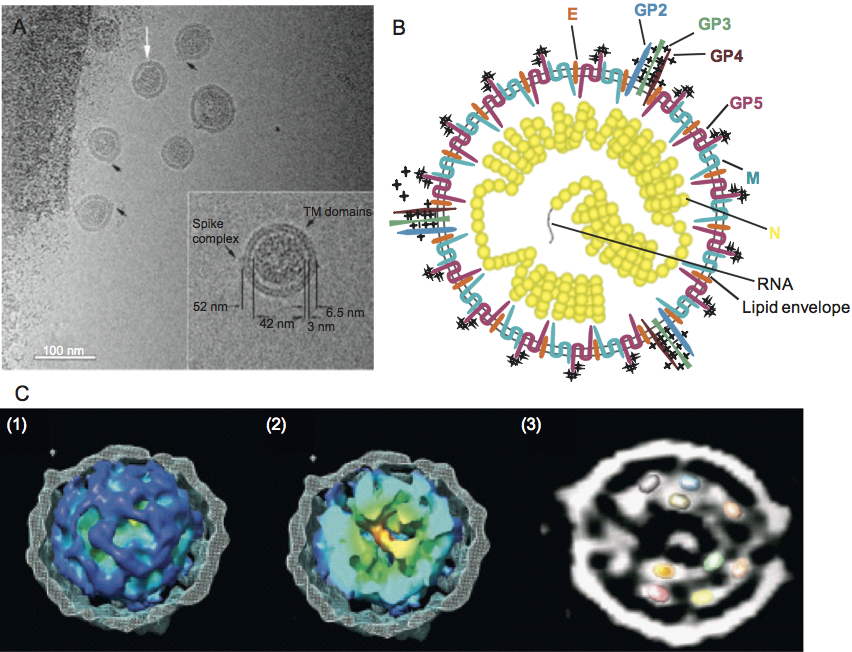
Figure 2 Arterivirus genome organization and expression. The general genome organization is shown at the top of the figure with the ORFs represented. The proteins encoded by the ORFs are indicated, while ORF colors for virion-associated proteins match those used in Figure 1B. Other domains: L, 5 leader sequence or 5 UTR; 3 UTR and a 3 poly(A) tail (zigzag line); filled circle, ORF 1a and 1b ribosomal frameshift site. The grey boxes immediately below represent the regions where PRRSV, LDV, and SHFV contain major insertions compared to EAV. In the infected cell, the full-length genome uses discontinuous transcription during minus strand synthesis to eventually produce a nested set of subgenomic messages (sgRNA), shown below. The proteins produced are listed to the right of the respective RNA. The structural protein composition of SHFV except for GP5, M, and N is unknown at present.
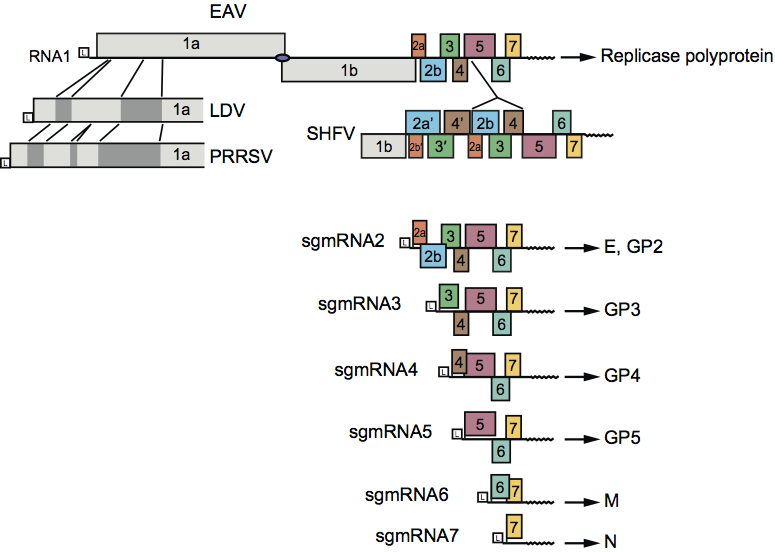
Figure 3 (A) Schematic of the major envelope proteins. The GP5 glycoproteins of LDV and PRRSV (left) and EAV (right) are disulfide bonded to the M protein. One neutralization domain has been identified on LDV and PRRSV, and 4 (A-D) on EAV. Hypervariable regions surround the neutralization domains. N-glycan addition (star) varies among arteriviruses, but the N-glycan residues closest to the disulfide bond between GP5 and M (S-S) are mostly conserved. (B) Predicted conformation of the minor envelope proteins in eukaryotic membranes.
(Reproduced with modifications from Perlman et al. (2008). ASM Press, Washington, DC with permission.)
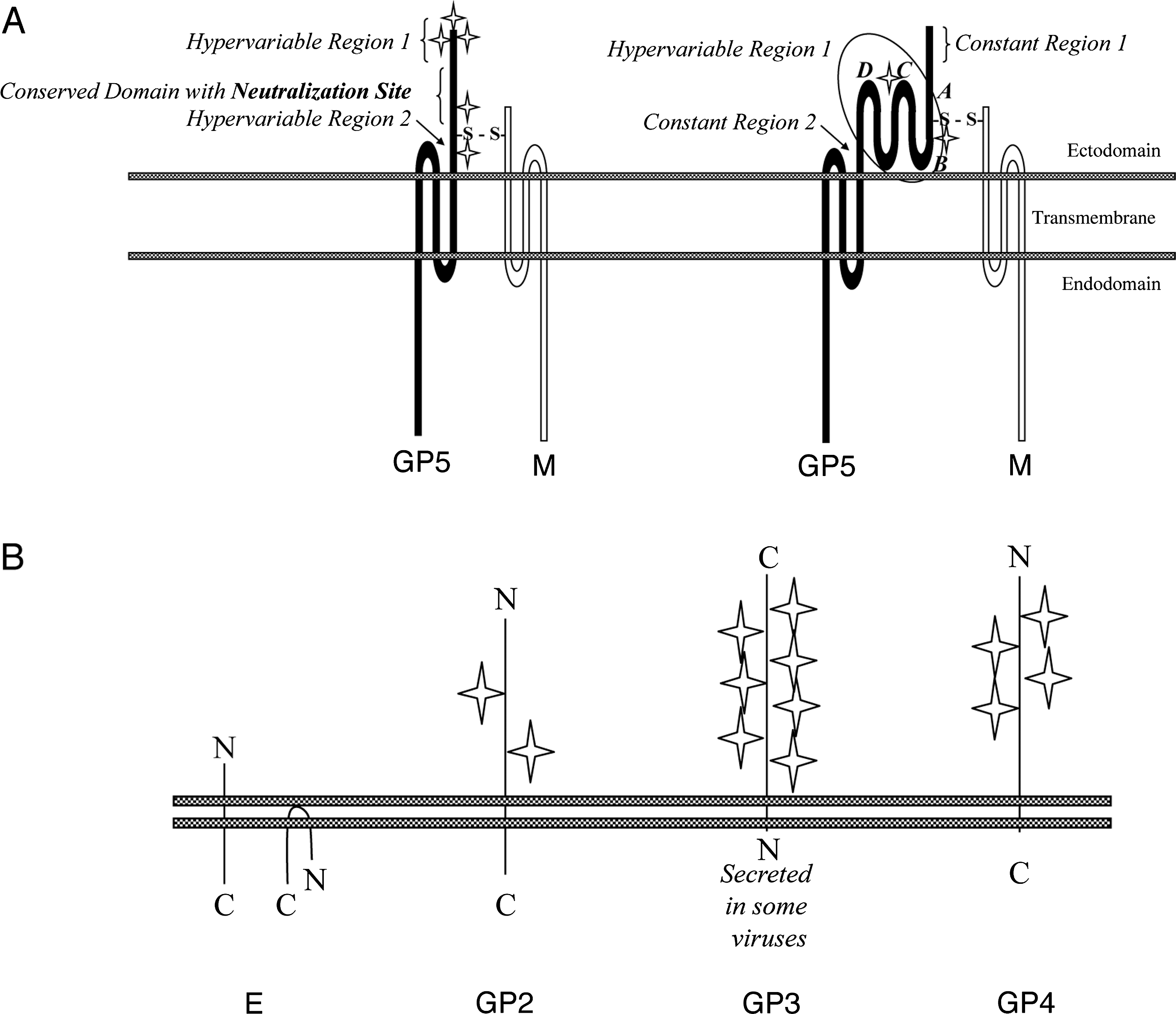
Figure 4 Overview of the replication cycle of the arterivirus prototype (EAV). The genome organization, including replicase cleavage sites (arrowheads; see also Figure 5), is shown at the top of the figure. Abbreviations: ER, endoplasmic reticulum; PM, plasma membrane; DMV, double membrane vesicle; NC, nucleocapsid.
(Modified from the Eighth ICTV Report.)
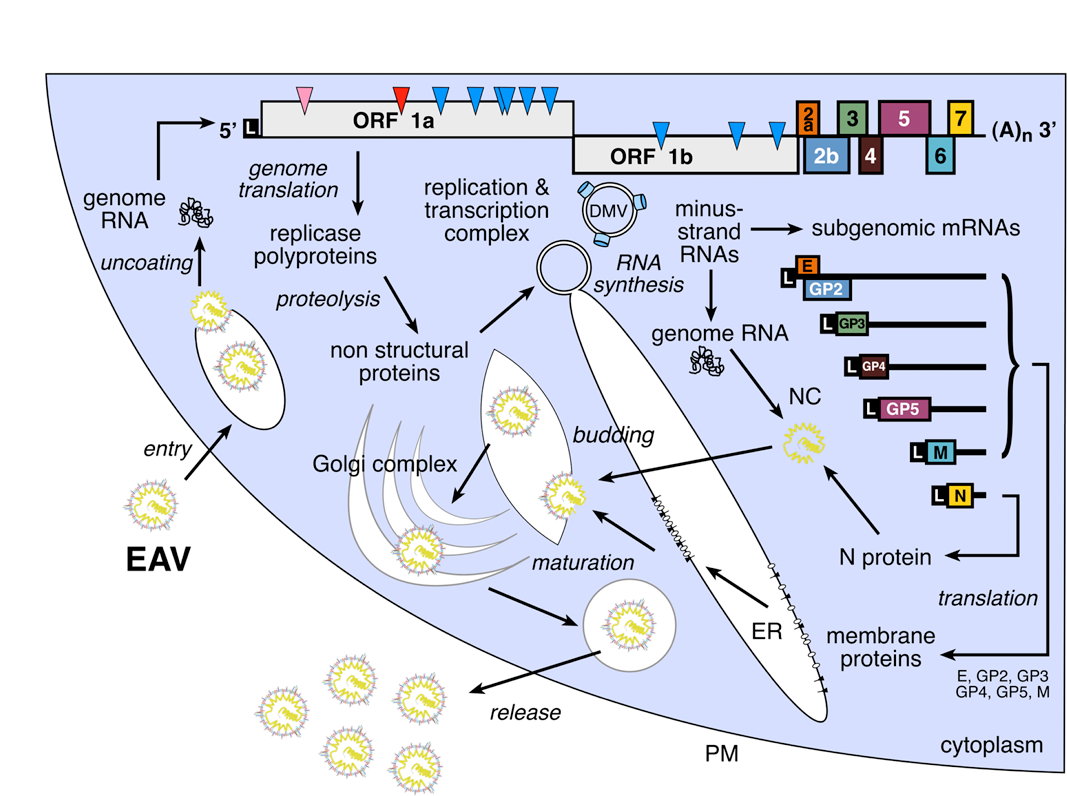
Figure 5 Overview of proteolytic processing of the EAV replicase polyproteins, with examined or potential similarities with PRRSV and LDV indicated. The domains for SHFV have not been described. Polyprotein cleavage sites are depicted with arrowheads matching the colour of the proteinase involved. Abbreviations: ZF, zinc-finger; 1 (PLP1), papain-like cysteine proteinase; 2 (PLP2), papain-like, cysteine proteinase; SP (also called Mpro), chymotrypsin-like serine proteinase; TM, transmembrane domain; RdRp, RNA-dependent RNA polymerase; Z, zinc binding; HEL, NTPase/helicase; U, nidoviral endonuclease specific for U (NendoU). In addition, several cysteine/histidine (C/H) recognized motifs, as well as the ribosomal frameshift site (RFS), are indicated below the figures. In EAV, PLP1 has become inactivated although its remnants were detected in the EAV nsp1 region. The large hypervariable region in PRRSV nsp2 that is characterized by insertions and deletions among different strains is shown by the hatched gray bar.
Figure 6 Phylogenetic analysis of the Arteriviridae was completed using the nucleotide sequences of SHFV (strain LVR 42-0; NC_003092), EAV (strain Bucyrus VBS; DQ846750), LDV-P (Plagemann strain; NC_001639), Type 1 PRRSV (strain Lelystad; M96262) and Type 2 PRRSV (strain VR-2332; U87392). Unrooted phylogenetic analysis of the full-length ORF1b replicase polyprotein. The tree was generated using the Dayhoff substitution model in PHYML, after alignment with the MUSCLE Alignment algorithm (default parameters, 8 iterations) in Geneious Pro 4.8.3 (Biomatters Ltd). Distance lengths represent substitutions per site. (K.S. Faaberg, unpublished data.)