Family: Rhabdoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are 100–430 nm in length and 45-100 nm in diameter. Defective virus particles are proportionally shorter. Viruses infecting vertebrates are bullet-shaped or cone-shaped; viruses infecting plants mostly appear bacilliform when fixed prior to negative staining; in unfixed preparations they may appear bullet-shaped or pleomorphic. The outer surface of virions (except for the quasi-planar end of bullet-shaped viruses) is covered with projections (peplomers) which are 5–10 nm long and about 3 nm in diameter. They consist of trimers of the viral glycoprotein (G). A honeycomb pattern of peplomers is observed on the surface of some viruses. Internally, the nucleocapsid, about 30–70 nm in diameter, exhibits helical symmetry and can be seen as cross-striations (spacing 4.5–5 nm) in negatively stained and thin-sectioned virions. The nucleocapsid consists of a ribonucleoprotein (RNP) complex comprising the genomic RNA and tightly bound nucleoprotein (N) together with an RNA-dependent RNA polymerase (L) and phosphoprotein (P). The RNP complex is active for transcription and replication: the N-RNA template is processed by the L protein, which contains most enzymatic activities, and its cofactor the P protein. In the cytoplasm, the RNP complex is uncoiled and filamentous, about 700 nm in length and 20 nm in diameter (Figure 1). In the virion, the lipid envelope containing the G protein interacts with the coiled RNP complex via the matrix protein (M).
Physicochemical and physical properties
Virion Mr is 300×106–1000×106 and S20w is 550–1045S (plant rhabdoviruses have larger S20,w values). Virion buoyant density in CsCl is 1.19–1.20 g cm−3; in sucrose it is 1.17–1.19 g cm−3. Virus infectivity is rapidly inactivated at 56 °C, or following UV-, gamma- or X-irradiation, or exposure to formalin or to lipid solvents such as detergents.
Nucleic acid
Virions contain a single molecule of linear, negative sense ssRNA (Mr 4.2×106–4.6×106, about 11–15 kb in size). The RNA represents about 1–2% of virion weight. The RNA has a 3′-terminal free hydroxyl group and a 5′-triphosphate and is not polyadenylated. The ends have inverted complementary sequences with transcription and replication initiation signals. Defective RNAs, usually significantly shorter than full-length RNA (less than half size), may be identified in RNA recovered from virus populations. They are usually negative sense; however, hairpin RNA forms are also found. Defective RNAs replicate only in the presence of homologous and, occasionally, certain heterologous helper rhabdoviruses which provide the functional genes. Full-length positive sense RNA, which is an intermediate during the replication process, may constitute up to 5% of a viral RNA population. Like the full-length negative sense RNA genome, it is permanently bound to N protein.
Proteins
Viruses generally have five structural polypeptides (designated L, G, N, P and M; see Table 1 for summary of their location, sizes and functions). The functions of other proteins, including additional non-structural glycoproteins (in ephemeroviruses) or C proteins (in a different ORF of the P mRNA for vesiculoviruses and lyssaviruses) are not known. Plant-adapted viruses have one or more additional non-structural proteins, one of which is thought to facilitate virus movement between plant cells. The structural proteins represent 65–75% of the virus dry weight. For certain viruses, other nomenclature has previously been used for the P protein (NS, M1 or M2) and the M protein (M1 or M2). RNA-dependent RNA polymerase, 5′ capping, guanosyl transferase, poly(A) polymerase, protein kinase, nucleoside triphosphatase and nucleoside diphosphate kinase activities are harboured by the nucleocapsid. Most catalytic functions have been attributed to L.
Table 1 Location and functions of rhabdovirus structural proteins
| Protein | Location, size and function |
| L | A component of the viral nucleocapsid (ca. 220–240 kDa) responsible for most of the functions required for transcription and replication: RdRp, mRNA 5′-capping, 3′-poly(A) synthesis and protein kinase activities. Observed sizes on SDS-PAGE are 150–190 kDa |
| G | Associated into trimers to form the virus surface peplomers (monomer ca. 65–90 kDa). Binds to host cell receptor(s), induces virus endocytosis then mediates fusion of viral and endosomal membranes. G is variously N-glycosylated and palmitoylated; it lacks O-linked glycans and has hemagglutinin activity. Induces and binds virus-neutralizing antibodies and elicits cell-mediated immune responses. G is involved in tropism and pathogenicity |
| N | Major component of the viral nucleocapsid (ca. 47–62 kDa). It associates with full-length negative and positive sense RNAs, or defective RNAs, but not mRNAs. N is not “inert” but an active element of the template, presenting the bases to the polymerase. Newly synthesized N probably modulates the balance between genome transcription and replication by influencing the recognition of the transcription signals. N elicits cell-mediated immune responses and humoral antibodies. In all nucleorhabdoviruses examined, N re-localises to a subnuclear compartment when co-expressed with the cognate P protein |
| P | A cofactor of the viral polymerase (ca. 20–30 kDa). It is variously phosphorylated and migrates on SDS-PAGE as a protein of about 40–50 kDa. The P of the nucleorhabdoviruses migrates faster. P protein is essential for at least two fundamental functions: (1) it mediates the physical link and the correct positioning of the RdRp (L) on the N-RNA template; (2) it acts as a chaperone during the synthesis of N, by forming N–P complexes that prevent N from self-aggregation and binding to cellular RNA. During the genome replication process, N is then transferred from these N–P complexes to the nascent viral RNA in order to ensure its specific encapsidation into new RNP. P elicits cell-mediated immune responses |
| M | A basic protein that is an inner component of the virion (ca. 20–30 kDa). It is believed to regulate genome RNA transcription. M binds to nucleocapsids and the cytoplasmic domain of G, thereby facilitating the process of budding. It is sometimes phosphorylated or palmitoylated. M is found in the nucleus and inhibits host cell transcription. It also mediates other pathological effects (cell rounding for VSIV, apoptosis for lyssaviruses, intracellular accumulation of the inner nuclear membrane for PYDV) |
Lipids
Virions are composed of about 15–25% lipids, with their composition reflecting that of the host cell membrane where virions bud. Generally phospholipids represent about 55–60%, and sterols and glycolipids about 35–40% of the total lipids. G protein may have covalently associated fatty acids proximal to the lipid envelope.
Carbohydrates
Virions are composed of about 3% carbohydrate by weight. The carbohydrates are present as N-linked glycan chains on G protein and as glycolipids. In mammalian cells, the oligosaccharide chains are generally of the complex type; in insect cells they are of non-complex types.
Genome organization and replication
Viruses contain at least five ORFs in the negative sense genome in the order 3′-N-P-M-G-L-5′ (e.g., for vesicular stomatitis Indiana virus; VSIV), or the equivalent. The corresponding cistrons are flanked by conserved start and stop transcription signals, about 10 nt in length. For certain viruses additional genes are interposed (Figure 2). Genes are transcribed processively (from 3′ to 5′ of the template virus RNA and in decreasing molar abundance) as 5′-capped, 3′-polyadenylated and generally monocistronic mRNAs (Figure 3). Polycistronic mRNAs have been identified for viruses in some species; they result from the readthrough or the absence of a stop transcription signal thereby allowing transcription extension across the adjacent 5′-cistron. A short uncapped, unpolyadenylated and untranslated leader RNA, corresponding to the complement of the 3′ terminus of the viral RNA (i.e., preceding the N mRNA), is also transcribed. Unlike mRNAs, leader RNA has a 5′ triphosphate terminus (Figure 3). Leader RNA of some viruses has been identified in the nucleus of infected cells. The mRNAs generally have common 5′-terminal sequences (e.g., m7Gppp(m)AmA(m)CA for vesiculoviruses and lyssaviruses) corresponding to the cap structure fused to the first nucleotides copied from the start transcription signal. The mRNAs also each contain a 3′-poly(A) tail which is produced by the viral transcriptase upon copying in a reiterative mode the 7 U residues terminating each stop transcription signal. Intergenic sequences are generally short but may be up to about 100 nt in length. In certain cases the 5′ end of an mRNA overlaps the 3′ end of the preceding gene.
Except for plant rhabdoviruses, which generally penetrate the cell through mechanical damage caused by insect vectors, rhabdovirus adsorption is mediated by G protein attachment to cell surface receptors and penetration of the cell is by endocytosis via coated pits. Various candidate receptors have been postulated for rabies virus (RABV) (nicotinic acetylcholine receptor AChR, neural cell adhesion molecule NCAM, low affinity nerve growth factor receptor p75NTR), vesicular stomatitis viruses (VSV) (phosphatidyl serine), viral hemorrhagic septicemia virus (VHSV) (fibronectin), and others. In addition, carbohydrate moieties, phospholipids and gangliosides may play a complementary role for virus binding. After penetration by endocytosis, low pH within the endosome provokes fusion between endosomal and viral membranes, liberating the RNP complex into the cytoplasm. The pH-induced fusion depends on conformational changes of the glycoprotein, a process that is reversible upon raising the pH. Once the nucleocapsid is released into the cytoplasm, the genome RNA is repetitively transcribed (primary transcription) by the virion transcriptase. N protein removal does not occur since the transcriptase only recognizes the RNA-N protein complex as template. The capped and polyadenylated mRNAs are generally translated in cytoplasmic polysomes except for the G mRNA which is translated on membrane-bound polysomes. Transcription occurs in the presence of protein synthesis inhibitors indicating that it does not depend on de novo host protein synthesis. Following translation, RNA replication occurs in the cytoplasm (full-length positive sense and then full-length negative sense RNA synthesis).
Nucleorhabdoviruses replicate in viroplasms in the cell nucleus. Replication again occurs on the RNA-N protein complex and requires the newly synthesized N, P and L protein species to concomitantly encapsidate the nascent RNA into a nucleocapsid structure. Apart from freshly translated N-P-L proteins, replication may require host factors. However, vesiculoviruses can replicate in enucleated cells, indicating that newly synthesized host gene products are not required. It has been proposed that the concomitant binding of N protein to the nascent positive or negative sense viral RNA species may promote replication rather than transcription, by favoring readthrough of transcription termination signals. Replication leads to the synthesis of a full length positive sense antigenome RNA. This, in turn, serves as a replicative intermediate for the synthesis of negative sense genome RNA for the progeny virions. Following replication, further rounds of transcription (secondary transcription), translation and replication ensue. A typical feature of negative sense RNA viruses (shared by all members of the order Mononegavirales) is that the RNA genome (or antigenome) is never “naked” in the cell but is always encapsidated by the nucleoprotein. This RNA-N complex is the true template recognized by the viral polymerase (transcriptase or replicase).
Post-translational trafficking and modification of G protein involves translocation across the membrane of the endoplasmic reticulum, removal of the amino-proximal signal sequence and step-wise glycosylation in compartments of the Golgi apparatus. Depending on the cell, the G protein may move to the plasma membrane, particularly to the basolateral surfaces of polarized cells.
Viral nucleocapsid structures are assembled in association with M and lipid envelopes containing viral G protein. The site of formation of particles depends on the virus and host cell. For vesiculoviruses, lyssaviruses, ephemeroviruses and novirhabdoviruses, nucleocapsids are synthesized in the cytoplasm and viruses bud from the plasma membrane in most, but not all cells. Some lyssaviruses bud predominantly from intracytoplasmic membranes and in some cases prominent virus-specific cytoplasmic inclusion bodies containing N protein are observed in infected cells (RABV inclusion bodies are called Negri bodies). Cytorhabdoviruses bud from intracytoplasmic membranes associated with viroplasms; none has been observed to bud from plasma membranes. Nucleorhabdoviruses bud from the inner nuclear membrane and accumulate in the perinuclear space.
Depending on the virus and host cell type, virus infections may inhibit cellular syntheses and cause apoptosis. The mechanisms are under investigation. Complementation between viral mutants of related viruses may occur (e.g., between vesiculoviruses), but not between viruses representing distinct genera. Complementation is also reported to occur by re-utilization of the structural components of UV-irradiated virus (VSIV). Inter-molecular genetic recombination between different virus isolates is very rare, but intra-molecular recombination may occur during the formation of defective RNAs. Phenotypic mixing occurs between some animal rhabdoviruses and other enveloped animal viruses (e.g., paramyxoviruses, orthomyxoviruses, retroviruses, herpesviruses).
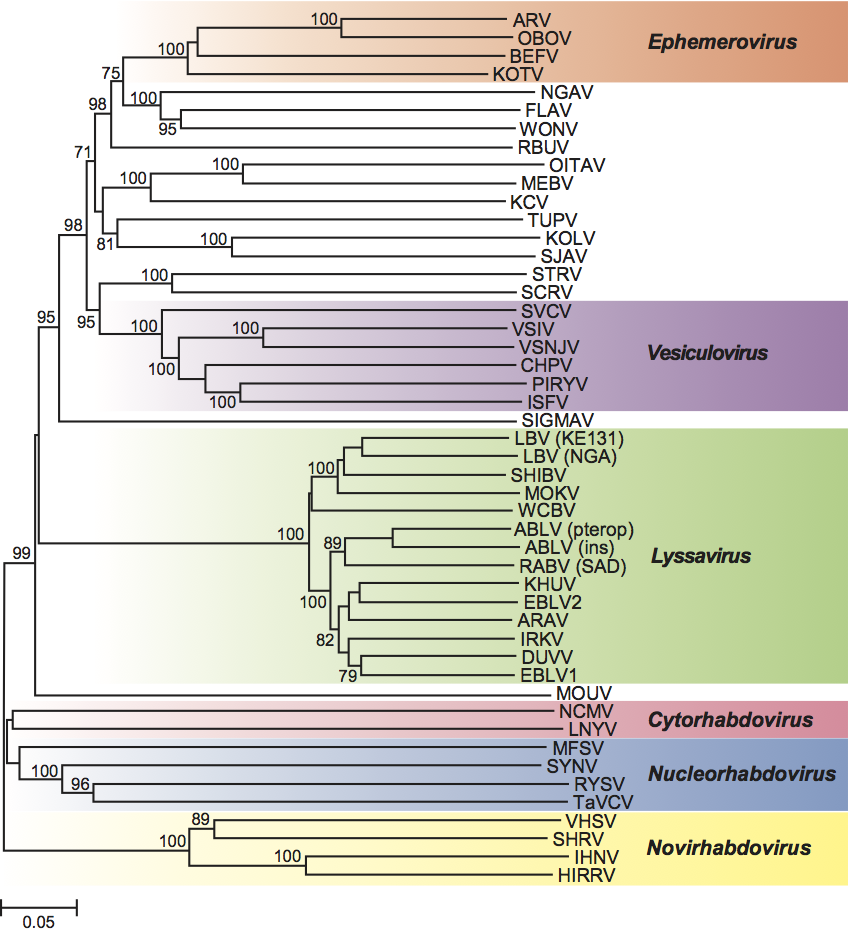
Six genera have been established, on the basis of significant differences in antigenicity, genome organization, replication sites and ecological properties (such as host range, pathobiology and circulation patterns). Phylogenetic relationships based on nucleotide and protein sequences support assignments of species to the identified genera.
Biological properties
Some rhabdoviruses replicate only in mammals, or birds, or fish, or arthropods, or other invertebrates, many have both arthropod and vertebrate hosts (arboviruses), while some species infect plants and certain plant-feeding arthropods. Some of the vertebrate rhabdoviruses have a wide experimental host range. A diverse range of vertebrate and invertebrate cell lines are susceptible to vertebrate rhabdoviruses in vitro. The viruses of plants usually have a narrow host range among higher plants; they replicate in insects and some replicate in insect cell cultures.
Sigma virus was recognized first as a congenital infection in drosophila. No rhabdovirus is known to be transmitted vertically in vertebrates or plants, but transovarial transmission has been documented in insects. Vector transmission may involve mosquitoes, sandflies, culicoids, aphids, leafhoppers or planthoppers. Some viruses are transmitted mechanically in sap or from the body fluids of infected hosts. Mechanical transmission of viruses infecting vertebrates may be by contact, aerosol, bite, or venereal.
Genus Vesiculovirus
Type species Vesicular stomatitis Indiana virus
Distinguishing features
Vesiculoviruses infect mammals, fish and insects. They have bullet-shaped virions and possess the shortest (11.0–11.3 kb) and simplest basic genome structure of all the rhabdoviruses. Five viral proteins are encoded in the following order from the 3′ genomic end: N, P, M, G and L. There is a small (47–50 nt) leader (l) sequence transcribed from the 3′ end and a 57–60 nucleotide trailer sequence at the 5′ end shown to be involved in viral replication. Most members of the genus encode two small highly basic proteins (C and C9) in a second ORF within the P protein. The function of these proteins remains undefined, as mutant viruses lacking them replicate normally in vitro. Interestingly, these ORFs are absent from the genome of Alagoas virus (VSAV). There are approximately 70 non-transcribed nucleotides in the vesiculovirus genome, two nucleotides at each of the four gene junctions, three nucleotides at the l -N junction and the 60 nucleotide trailer sequence at the 5′ terminus. The four non-coding gene junctions separating the five mRNAs contain a 23 nucleotide conserved sequence: 3′-AUACUUUUUUUNNUUGUCNNUAG-5′ which functions as polyadenylation signal terminating each cistron, signals the start of the next mRNA species and templates the capped mRNA 5′ end (m7Gppp(m)Am-A(m)CAGNNAUC…).
Virion properties
Morphology
Virions are enveloped and bullet-shaped. Their three-dimensional structures have been recently determined utilizing cryo-electron microscopy (Figure 4). The nucleocapsid forms a conical-shaped tip and a helical structure in the trunk. Each virion contains an outer helix of matrix protein M and an inner helix of N protein and RNA. M has a hub domain with four contact sites which link to neighboring M and N subunits, providing rigidity by clamping adjacent turns of the RNP. Interactions among the N and M proteins provide scaffolding and stability to the complex and are critical in maintaining the bullet shape. Two layers of lipids obtained from the host cell membrane with embedded trimers of the envelope glycoprotein (G) form the outer layers of the virion and mediate the interaction with cellular receptor(s).
Nucleic acid
Viruses contain a single molecule of linear, negative sense ssRNA of about 11 kb in size. Defective RNAs, an artifact shown to occur during tissue culture passage of some vesiculoviruses, are usually significantly shorter than full-length RNA and can be found packaged into small virions known as defective interfering (DI) particles.
Proteins
Vesiculoviruses have five structural proteins (designated N, P, M, G and L). Most vesiculoviruses also have two small, highly basic proteins coded in a second ORF within P (termed C9 and C). The structural proteins represent 65–75% of the virus dry weight. For VSIV the numbers of molecules per infectious virus particle is estimated as: L (20–50); G (500–1,500); N (1,000–2,000); P (100–300); and M (1,500–4,000). Three viral proteins N, P and L combine to form the transcription and replication complex which has RNA-dependent-RNA polymerase (RdRP) activity. The N protein encapsidates the viral RNA and functions in close association with the P protein. The P protein is a highly phosphorylated protein associated with viral polymerase activity. It mediates the binding of the L protein to the nucleocapsid core and facilitates access of the polymerase to the RNA template during transcription and replication. Phosphorylation of P protein seems to be necessary for optimal transcriptase activity. The exact role of the nonstructural C and C9 proteins is unclear. Engineered viruses that do not express C proteins are indistinguishable from wild type virus in protein synthesis, virus production and host-protein synthesis shut off in tissue culture cells. The large L protein is multifunctional and performs most of the polymerase-associated functions including RNA synthesis, capping, methylation and poly (A) addition. It also has protein kinase activity which preferentially phosphorylates serine residues on the P protein. The matrix protein (M) is the most abundant protein of virions. M binds specifically to G protein monomers and promotes their trimerization. It also associates with N, provides rigidity and stability and is critical in maintaining the bullet shape.
The G protein is a typical class I membrane associated glycoprotein, with approximately 90% of the N-terminal region of the molecule projecting from the surface of the virion or infected cell, a hydrophobic transmembrane domain anchoring the protein in the membrane, and a C-terminal 28 amino acid cytoplasmic domain projecting to the interior of the infected cells. The G protein forms trimers that constitute the approximately 400 spikes that are embedded on the virion bilayer phospholipid envelope. The G protein plays a major role in attachment and penetration of vesiculoviruses into susceptible cells and budding of virions from infected cells. It is the major target of serotype-specific neutralizing antibodies and is capable of inducing cell membrane fusion at low pH.
Genome organization and replication
Total genomic RNA lengths range from 11,003 nt in Cocal virus (COCV) to 11,336 nt in field isolates of VSIV. The genome organization consists of a 47 nt leader sequence followed by the five structural protein genes in the order 3′ N-P-M-G and L and a 57–59 nt trailer sequence at the 5′ end. Most differences in length are found in non-translated regions, particularly in the M and G mRNAs. Despite the variability in lengths of the five mRNAs of the Indiana subtypes, four of the five predicted structural proteins (N, M, G and L) are within one amino acid residue in length of each other.
Table. Genome and gene length variation among vesiculoviruses
| Virus | Genome nt | N aa | P aa | M aa | G aa | L aa |
| VSIV | 11161-11336 | 422 | 265 | 229 | 511 | 2109 |
| VSNJV | 11123 | 422 | 274 | 229 | 517 | 2109 |
| ISFV | 11088 | 423 | 289 | 226 | 523 | 2093 |
| SVCV | 11047 | 418 | 309 | 223 | 509 | 2101 |
| CHPV | 11119 | 422 | 293 | 229 | 530 | 2092 |
| PIRYV | N.D. | N.D. | 327 | 229 | 529 | N.D. |
| COCV | 11003 | 422 | 264 | 229 | 512 | 2108 |
| VSAV | 11070 | 422 | 260 | 229 | 511 | 2108 |
N.D. = not determined
Abbreviations are defined in Tables 2-16
Vesiculovirus replication is well studied and has served as model for replication of the rhabdoviruses. Transcription begins at a single entry site at the 3′ end of the genome with each gene expressed as a capped and polyadenylated monocistronic mRNA. The first transcript is the 3′ leader, which is neither capped nor polyadenylated. It is transported to the nucleus where it inhibits host cell transcription. The leader transcript is followed by the N mRNA, which is capped during synthesis by the virion polymerase complex (composed of N, P and L). The intergenic sequence (5′-AGUUUUUUUCAUA-3′) signals polyadenylation, termination and re-initiation of transcription in decreasing amounts as the polymerase complex moves away from the single entry site. Therefore, the gene order (i.e. 3′ N>P>M>G>L 5′) provides an efficient way of regulating gene expression, by which proteins necessary in larger amounts (such as N) are located near the 3′ end and are transcribed in larger amounts and those needed in smaller amounts are located towards the 5′ end and are transcribed less frequently.
Following translation of the mRNAs to yield the viral proteins, genome replication starts. In the replication process the RdRP initiates at the 3′ end of the genome, ignores all the signals for stop and polyadenylation of individual mRNAs and instead synthesizes a full-length complementary antigenome. This in turn serves as template for synthesis of a 45-nucleotide minus sense leader RNA, and also for synthesis of full-length progeny genomes. Full-length genomes can either serve as templates for secondary transcription, or can be assembled into infectious particles. Factors determining the RdRP functions in transcription or replication modes are not fully understood but the proportion of N protein and available RNP templates are thought to be critical for determining these functions. Following replication, further rounds of transcription (secondary transcription), translation and replication ensue.
Antigenic properties
Vesiculoviruses have been classified into serotypes based on their neutralization pattern which is determined by epitopes located in the viral G protein. The G protein also determines protection in animals vaccinated either with inactivated whole virus or with subunit vaccines. Furthermore, recombinant viruses containing the G protein of vesicular stomatitis New Jersey virus (VSNJV), but all other proteins from VSIV, induced protective immune responses against challenge with VSNJV. The N protein is a cross-reactive antigen used in complement fixation tests that help define members of the genus. Weak serological cross-reactions may occur between viruses in different genera. One of the criteria used for vesiculovirus classification is cross-reactivity by complement fixation.
In the case of vesicular stomatitis viruses there are two major serotypes: New Jersey (VSNJV) and Indiana. The serotype Indiana has been subdivided into three distinct serological complexes. Indiana 1 comprises the classical Indiana viruses (VSIV) found from Northern South America to Southern US. The Indiana 2 subtype has (COCV) as the prototype virus which was originally isolated from mites collected from rice rats in Trinidad in 1961. Indiana 2 viruses cause disease in cattle and horses in Brazil and Argentina. The Indiana 3 subtype is represented by (VSAV) which was first isolated from a mule in Alagoas, Brazil, in 1964. This subtype is the most common cause of vesicular stomatitis in livestock in Brazil. There are other vesiculoviruses that infect mammals, some cause disease in humans, e.g. (Piry virus, Isfahan virus and Chandipura virus) and some have been isolated from blood sucking insects but to date are not known to cause disease in vertebrates (e.g., Maraba virus and Carajas virus).
In the case of the fish vesiculovirus Spring viremia of carp virus (SVCV) there is a single serotype. Antibodies directed against SVCV cross-react to various degrees with other fish rhabdoviruses; pike fry rhabdovirus (PFRV), grass carp virus (GrCRV) and tench rhabdovirus (TenRV), indicating that the viruses are closely related. SVCV and PFRV have been shown to share common antigenic determinants on the G, N and M proteins, but can be differentiated by neutralisation assays.
Biological properties
Vesiculoviruses cause disease in mammals or fish. Those causing disease in mammals are transmitted by insects and therefore are considered arboviruses. The natural cycle of vesiculoviruses infecting mammals remains largely unknown but these viruses are commonly found in insects of a number of species and serological evidence suggests they are capable of infecting not only a number of wild mammals, but also birds and even reptiles living in endemic areas. This wide range of hosts might explain the ability of vesiculoviruses to infect and replicate in a very diverse range of vertebrate and invertebrate cells in vitro. In fact, mammalian vesiculoviruses are able to be transmitted both by insects and by contact. Vesicular stomatitis viruses are transmitted to cattle, horses and pigs by various blood-sucking insects found to be infected during epidemics, including sandflies, blackflies and culicoids and also can be transmitted between mammals by direct contact. Experimental mechanical transmission also has been achieved by feeding vesiculovirus laboratory infected grasshoppers to cattle. However, grasshoppers have never been shown to carry vesiculoviruses in nature. Vesiculoviruses have been shown to be transmitted not only transovarially but, interestingly, also horizontally between infected and non-infected black-flies while co-feeding on mammalian hosts. The latter means of transmission might explain the noticeable absence of viremic mammal hosts for vesiculoviruses, an unusual feature for an arbovirus.
In the case of SVCV the hosts are predominantly cyprinid fish. Naturally occurring SVC infections have been recorded from common carp (Cyprinus carpio carpio) and koi carp (Cyprinus carpio koi), crucian carp (Carassius carassius), sheatfish (also known as European catfish or wels) (Silurus glanis), silver carp (Hypophthalmichthys molitrix), bighead carp (Aristichthys nobilis), grass carp (white amur) (Ctenopharyngodon idella), goldfish (Carassius auratus), orfe (Leuciscus idus) and tench (Tinca tinca). The virus can be transmitted by ectoparasites such as carp lice (Argulus foliaceus) and the leeches (Pisicola geometra), but waterborne transmission without any vector organism is also effective.
The replication temperature range of SVCV is typically lower than those of the mammalian rhabdoviruses, reflecting the aquatic poikilothermic nature of the host species, and the viruses are typically isolated on cultured fish cell lines at 15–25 °C. The disease patterns are influenced by water temperature, age and condition of the fish, population density and stress factors. The immune status of the fish is also an important factor with both non-specific (interferon) and specific immunity (serum antibodies, cellular immunity) having important roles. Clinical disease is usually observed at water temperature between 5–18 °C and is most severe at temperatures below 10 °C, when it is believed the host immune response is suppressed or delayed.
Species demarcation criteria in the genus
Vesiculovirus species have been defined primarily by serological means coupled with phylogenetic analysis of the genomes. Biological characteristics such as host range and mechanisms of transmission are also used to distinguish viral species within the genus.
List of species in the genus Vesiculovirus
| Carajas virus |
|
|
| Carajas virus | [FW339542] | (CJSV) |
| Chandipura virus |
|
|
| Chandipura virus CIN0451 | [GU212856] | (CHPV-CIN0451) |
| Cocal virus |
|
|
| Cocal virus Indiana 2 | [EU373657] | (COCV-Ind2) |
| Isfahan virus |
|
|
| Isfahan virus | [AJ810084] | (ISFV) |
| Maraba virus |
|
|
| Maraba virus |
| (MARAV) |
| Piry virus |
|
|
| Piry virus | [Z15093*, D26175*] | (PIRYV) |
| Spring viraemia of carp virus |
|
|
| Spring viraemia of carp virus VR-1390 | [AJ318079] | (SVCV-VR1390) |
| Vesicular stomatitis Alagoas virus |
|
|
| Vesicular stomatitis Alagoas virus Indiana 3 | [EU373658] | (VSAV-Ind3) |
| Vesicular stomatitis Indiana virus |
|
|
| Vesicular stomatitis Indiana virus 98COE | [AF473864] | (VSIV-98COE) |
| Vesicular stomatitis New Jersey virus |
|
|
| Vesicular stomatitis New Jersey virus | [K02379*, S61075*, J02433*, M20166*, M14553*, K02747*] | (VSNJV) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Vesiculovirus but have not been approved as species
| BeAn 157575 virus |
| (BeAnV-157575) |
| Boteke virus | [GU816014*] | (BTKV) |
| Calchaqui virus |
| (CQIV) |
| Eel virus American |
| (EVA) |
| Eel virus European X | [FN557213] | (EXEV) |
| Grass carp rhabdovirus |
| (GrCRV) |
| Gray Lodge virus |
| (GLOV) |
| Jurona virus | [GU816024*] | (JURV) |
| Klamath virus |
| (KLAV) |
| Kwatta virus |
| (KWAV) |
| La Joya virus |
| (LJV) |
| Malpais Spring virus |
| (MSPV) |
| Perinet virus | [AY854652*] | (PERV) |
| Pike fry rhabdovirus | [FJ872827] | (PFRV) |
| Porton virus | [GU816013*] | (PORV) |
| Radi virus |
| (RADIV) |
| Tench rhabdovirus |
| (TenRV) |
| Ulcerative disease rhabdovirus |
| (UDRV) |
| Yug Bogdanovac virus |
| (YBV) |
* Sequences do not comprise the complete genome.
Genus Lyssavirus
Type species Rabies virus
Distinguishing features
Lyssaviruses share common genome organization and constitute a well-delineated monophyletic group within the family Rhabdoviridae. Genetic distances between lyssavirus species are significantly shorter than the distances between viruses in other rhabdovirus genera, which has been attributed to evolutionary constraints, possibly imposed by their unique pathobiology or their vectors/reservoirs preference. These viruses cause acute progressive encephalitis (rabies) in mammals, being transmitted between susceptible individuals directly by bites, scratches or contamination of mucous membranes with saliva, without participation of arthropod vectors. Bats (order Chiroptera) are the principal reservoir hosts for the majority of lyssaviruses, whereas “terrestrial” carnivores (order Carnivora), as well as bats, maintain circulation of rabies virus (RABV). Viruses of the genus are distributed worldwide, except Antarctica and several isolated islands, although viruses of different species have different circulation ranges. Based on phylogenetic relationships and antigenic properties, the genus has been subdivided into two phylogroups. Phylogroup I includes RABV, Australian bat lyssavirus (ABLV), Duvenhage virus (DUVV), European bat lyssaviruses 1 and 2 (EBLV1 and 2), Aravan virus (ARAV), Khujand virus (KHUV) and Irkut virus (IRKV), whereas phylogroup II includes Lagos bat virus (LBV) and Mokola virus (MOKV). The most divergent species in the genus, West Caucasian bat virus (WCBV), is not a member of either of these phylogroups.
Virion properties
Morphology
The virions are bullet-shaped, 60–110×130–250 nm in size, and composed of two structural units: an internal helical nucleocapsid, about 50 nm in diameter, and a lipid envelope which is derived from the host cytoplasmic membrane during budding. The nucleocapsid is comprised of a ribonucleoprotein (RNP) complex consisting of RNA genome, nucleoprotein (N), phosphoprotein (P) and RNA-dependent RNA polymerase (L). The exact position of the matrix protein (M) remains controversial, and may be either contained in the central channel of the RNP or form a helix between RNP and virion membrane, as has been shown for vesiculoviruses. The glycoprotein (G) is composed of four distinct domains: a signal peptide (non-structural), ectodomain, transmembrane peptide and cytoplasmic domain. Knobbed spikes (10 nm in length), consisting of three glycosylated ecto-domains, protrude through the virion membrane.
Physicochemical and physical properties
Lyssaviruses such as RABV consist of RNA (2–3%), protein (67–74%), lipid (20–26%) and carbohydrate (3%). The five major polypeptides include: N (58–62 kDa), P (35–40 kDa), M (22–25 kDa), G (65–80 kDa), and L (190 kDa). The G protein of RABV is glycosylated and palmitoylated at sites that have been mapped. N and P are both phosphoproteins and in the case of RABV, phosphorylation has been shown to involve several different host protein kinases: for N the cellular casein kinase II is implicated; P is phosphorylated by certain isomers of protein kinase C as well as by an additional kinase, RVPK, yet to be clearly defined. In addition a viral encoded protein kinase activity has been postulated in the L protein. The N:P ratio in the RNP of RABV is 2:1 per virion, which indicates that two molecules of N interact with 1 molecule of P. The M is not phosphorylated, it collaborates with both RNP and the G. The G is the only glycosylated protein, with branched chain oligosaccharides, which account for 10–12% of the total mass of the protein. The lyssavirus virion envelope contains other host-derived minor protein components. A lipoprotein bilayer consists of a mixture of host-derived lipids, including phospholipids, neutral lipids and glycolipids.
Nucleic acid
The negative sense ssRNA is about 12 kb in length. The RNA is tightly associated with the N protein within the RNP.
Genome organization and replication
The genome consists of a leader sequence (ca. 50 nt), followed by five structural protein genes in the order 3′-N-P-M-G-L-5′, separated by non-transcribed intergenic regions and followed by an ~70 nt trailer. Transcription initiation signal of each mRNA is conserved (3′-UUGURRNGA-5′), as well as the transcription termination-polyadenylation signal (3′-UCUUUUUUUG-5′). Untranslated regions of mRNAs are relatively short, except the 3′UTR of the G mRNA, which is about 400–700 nucleotides in length. Intergenic regions are variable (2–100 nucleotides), but their lengths increase along the 3′–5′ direction, which has the potential effect of causing a progressive slowing and decreasing efficiency of transcription.
Transcription and replication occur in the cytoplasm of infected cells. After attachment to receptors of the host cell membrane, mediated by the glycoprotein spikes, the virus enters the cell via the endosomal pathway. The pH decrease within the endosome leads to G protein-mediated fusion between endosomal and viral membranes and the RNP is liberated into the cytoplasm. Further, monocistronic positive sense RNAs, corresponding to the leader RNA and the five mRNAs, are transcribed in cascade from the genome encapsidated by the N protein. Transcription is mediated by the viral RNA-dependent RNA polymerase (L) and its co-factor, the P protein. All but one of the monocistronic mRNAs produce a single protein from a single ORF initiated at the first AUG initiation codon. However, the P mRNA yields three or four proteins, initiated from secondary downstream in-frame AUG codons.
As translation products of the most proximal (i.e. abundant) mRNAs during transcription, N and P proteins are produced in greater quantity within the cytoplasm. P acts as a chaperone during the synthesis of N, by forming N–P complexes which keep N soluble while preventing it from binding to cellular RNA. N is then transferred from these N–P complexes to the nascent viral RNA in order to ensure its specific encapsidation. Some of the N protein encapsidates the leader transcript. A switch from transcription to replication produces full-length complementary (positive sense) antigenome copies, which then become the template for progeny negative sense genome RNA. Both antigenome and genome RNAs are co-transcriptionally encapsidated by the soluble N and form RNPs to protect the RNA from enzymatic degradation. Both contain a specific 5′-terminal cis-acting encapsidation signal which is first recognized by N, then encapsidation proceeds in a cooperative way in the 5′–3′ direction.
Antigenic properties
Relatively short genetic distances between lyssavirus species correlate with their antigenic cross-reactivity. Antigens of RNP, which are most abundant in infected cells, cross-react between all members of the genus described to date. This feature facilitates the use of standardized diagnostic reagents for detection of all lyssaviruses (for example, by direct fluorescent antibody or immunohistochemical assays). In contrast, outer glycoprotein antigens are relatively conserved within phylogroups (ectodomain conservation >75%) but not between phylogroups (ectodomain conservation <65%). As a result, commercially available rabies vaccines and anti-rabies immunoglobulins, that mainly induce or provide neutralizing antibodies targeting the glycoprotein, provide protection against phylogroup I lyssaviruses but not against LBV, MOKV and WCBV.
Biological properties
Lyssaviruses are essential neurotropic pathogens. Delivered into a wound via a bite or wound contamination, the virus can replicate at the inoculation site, as was shown for skeletal muscle cells. Then the virus reaches the motor or sensory neurons and propagates up to the central nervous system (CNS) by following neuronal connections and using retrograde axonal transport. Neuronal pathways shield the virus from host immune surveillance, resulting in absence of early antibody response. Being delivered to the CNS, the virus disseminates rapidly. Nearly all regions of the CNS may be affected and RABV has been used to map neuronal connections. The duration of the asymptomatic incubation period might be variable (two months in average), while the symptomatic clinical period is rapid and severe (about one week). Neuropathological changes observed in the infected brain are relatively mild histologically and include gliosis, slight neuronophagia, perivascular infiltration with inflammatory cells, with rare involvement of meninges. Occasionally, more severe brain damage occurs, such as spongiform lesions, extensive neuronal degeneration and widespread inflammation. Functional alteration of the CNS is much more significant than is morphological representation. Reverse dissemination of virus from the CNS during the clinical period of rabies occurs along peripheral nerves. Viral RNA may be detected in a variety of organs and tissues at the end of the clinical period. However, only low titres of infectious virus occasionally can be isolated from extraneural tissues. The exception is the salivary glands, in which virus passes additional replication cycles and is released into the saliva to complete the transmission. Acute generalized CNS dysfunction leads to a lethal outcome of the disease. Very rare cases of survival after manifestation of clinical signs of rabies have occasionally been observed in humans and some animals. However, these sporadic events cannot be taken as support for a theory of lyssavirus persistence.
In nature, lyssaviruses are associated with particular mammalian reservoir hosts. Spill-over infections into vertebrates of other species typically lead to a dead end of the transmission chain in the vast majority of cases. The exception is RABV, which is distributed most broadly, and host shifts of certain variants with the establishment of sustained circulation in a new host of a different species have been well documented. Laboratory rodents (mice, hamsters) are highly susceptible to intra-cranial inoculation with lyssaviruses, whereas their susceptibility to peripheral inoculation varies, depending on viral species and lineage, inoculation dose and route. Cell cultures derived from the mammalian nervous tissue (such as mouse neuroblastoma, MNA cells) are more susceptible to lyssaviruses than are other mammalian cell cultures (BHK, Vero etc.). Successful propagation in insect cell lines has been described only for MOKV.
Species demarcation criteria in the genus
Until 1956, RABV was believed to be antigenically unique. The discovery of “rabies-related” viruses in Africa warranted the creation of the genus Lyssavirus (Greek lyssa: rage). The genus was at first divided into four serotypes by antigenic cross-reactivity with sera and monoclonal antibodies: RABV, LBV, MOKV and DUVV. Further isolations of new lyssaviruses in Europe, then Australia and Asia, and the progress in genetic characterization supported the delineation of lyssavirus genotypes, confirming and expanding antigenic data. Subsequent evaluation of long genome areas in various phylogenetic models, as well as recognition of additional characters outlined below, facilitated the classification of lyssaviruses into species.
In general, demarcation criteria for lyssavirus species include:
- Genetic distances, with the threshold of 80–82% nucleotide identity for the complete N gene, that provides a better quantitative resolution compared to other genes, or 80–81% nucleotide identity for concatenated coding regions of N+P+M+G+L genes. Globally, all isolates belonging to the same species have higher identity values than the threshold, except the viruses currently included into the LBV species. For that reason some authors suggested that LBV be subdivided into several genotypes. However, as these LBV representatives are segregated into a monophyletic cluster in the majority of phylogenetic reconstructions, in the absence of other sufficient demarcation characters there is currently no possibility to subdivide LBV into several viral species.
- Topology and consistency of phylogenetic trees, obtained with various evolutionary models.
- Antigenic patterns in reactions with anti-nucleocapsid monoclonal antibodies (preceded by serologic cross-reactivity and definition of lyssavirus serotypes, using polyclonal antisera).
- Whenever available, additional characters, such as ecological properties, host and geographic range, pathological features are recruited.
List of species in the genus Lyssavirus
| Aravan virus |
|
|
| Aravan virus – Kyrgyzstan | [EF614259] | (ARAV-KG) |
| Australian bat lyssavirus |
|
|
| Australian bat lyssavirus (pteropid bat variant) | [AF418014] | (ABLV-pb) |
| Australian bat lyssavirus (insectivorous bat variant) | [AF081020] | (ABLV-ib) |
| Duvenhage virus |
|
|
| Duvenhage virus 86132SA | [EU293119] | (DUVV-86132SA) |
| European bat lyssavirus 1 |
|
|
| European bat lyssavirus 1 8918FRA | [EU293112] | (EBLV1-8918FRA) |
| European bat lyssavirus 2 |
|
|
| European bat lyssavirus 2 9018HOL | [EU293114] | (EBLV2-9018HOL) |
| Irkut virus |
|
|
| Irkut virus - Russia | [EF614260] | (IRKV-RU) |
| Ozernoe | [FJ905105] |
|
| Khujand virus |
|
|
| Khujand virus - Tajikistan | [EF614261] | (KHUV-TJ) |
| Lagos bat virus |
|
|
| Lagos bat virus, lineage A | [EU293108] | (LBVSEN1985; 0406SEN) |
| Lagos bat virus, lineage B | [EU293110] | (LBVNIG1956; 8619NGA) |
| Lagos bat virus, lineage C | [EF547454*, EF547411*, EF547424*, EF547441*] | (LBVSA1980) |
| Lagos bat virus, lineage D | [GU170202] | (KE576) |
| Mokola virus |
|
|
| Mokola virus | [Y09762] |
|
| Rabies virus |
|
|
| Pasteur virus | [M13215] | (PV) |
| Street Alabama Dufferin, Bern-19 | [M31046] | (SAD B-19) |
| Silver-haired bat rabies virus | [AY705373] | (SHBRV-18) |
| Rabies virus (European fox) | [EU293115] | (9147FRA) |
| Rabies virus (south-east Asia, dog) | [EU293111] | (8764THA) |
| Rabies virus (North America, raccoon) | [EU311738] | (RRV ON-99-2) |
| Rabies virus (Argentina, free-tailed bat) | [EU293116] | (9704ARG) |
| West Caucasian bat virus |
|
|
| West Caucasian bat virus - Russia | [EF614258] | (WCBV-RU) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Lyssavirus but have not been approved as species
| Shimoni bat virus | [GU170201] | (SHIBV) |
Genus Ephemerovirus
Type species Bovine ephemeral fever virus
Distinguishing features
Ephemeroviruses are arthropod-borne animal rhabdoviruses. Bullet-shaped virions have a characteristic tapered appearance at the apical end and a prominent axial channel intruding from the base. The genome is large (>14 kb) and complex, containing a non-structural glycoprotein (GNS) gene and several small accessory protein genes. The GNS glycoprotein shares significant amino acid sequence identity with the virion glycoprotein (G) and appears to have arisen by gene duplication. Viruses in the genus cross-react strongly in complement-fixation (CF) and indirect immunofluorescence tests.
Virion properties
Morphology
Virions are bullet- or cone-shaped with a length of about 140–200 nm and diameter about 60–80 nm. They display a prominent axial channel intruding from the base and a precisely coiled, helical nucleocapsid with 35 cross-striations at intervals of 4.8 nm.
Physicochemical and physical properties
Virions have a buoyant density in CsCl of 1.19 g cm−3 and sedimentation coefficient of 625S. Viruses are sensitive to acid or alkali and most stable at pH 7.0–8.0. Bovine ephemeral fever virus (BEFV) particles contain at least five structural proteins, designated: L (180 kDa); G (81 kDa); N (52 kDa); P (43 kDa); and M (29 kDa). The G protein is a virus membrane-associated glycoprotein which contains five potential sites for attachment of N-linked glycans. The N protein is phosphorylated. The M protein is also phosphorylated in virions.
Nucleic acid
The genome comprises a single molecule of negative sense, single stranded RNA >14 kb.
Genome organization and replication
The 14.9 kb (−)ssRNA genome of BEFV contains 10 genes in the order 3′-N-P-M-G-GNS-α1-α2-β-γ-L-5′ and intergenic regions of between 26 and 53 nt. The γ and L genes overlap by 21 nt. Additional small ORFs occur in alternative frames in the P and α2 genes. Each gene, except α1, is initiated from a UUGUCC sequence (mRNA: 5′-cap-AACAGG…) and terminates at a putative polyadenylation site GNAC(U6-7)-3′. Each gene is transcribed as a monocistronic mRNA except the α1 and α2 genes which are transcribed in a bicistronic mRNA. The 15.0 kb Berrimah virus (BRMV) genome has the same gene order as BEFV and employs the same consensus transcription initiation and polyadenylation signals. However, unlike BEFV, intergenic regions are up to 100 nt in length, additional small ORFs do not occur in the P or α2 genes, and all genes appear to be transcribed independently. In ARV, the 14.6 kb genome contains nine genes in the order 3′-N-P-M-G-GNS-α1-α2-β-L-5′ and intergenic regions of 1–4 nt. The β and L genes overlap by 22 nt. ARV lacks a γ gene comparable to that of BEFV. An additional ORF occurs in an alternative frame in the P gene but not in the α2 gene. Each gene is initiated from a viral 3′-UUGUC sequence (mRNA: 5′-cap-AACAG…). However the putative polyadenylation signals are more variable than those of BEFV and may account for the synthesis of polycistronic mRNAs.
The GNS gene product is a 90 kDa non-virion glycoprotein which has been identified in BEFV-infected mammalian cells. GNS is highly glycosylated (eight potential sites for N-linked glycans). The G and GNS proteins, although not identical, exhibit homologies with each other and to lesser extents with the G proteins of other animal rhabdoviruses. In Adelaide River virus (ARV), the G protein (90 kDa) contains six potential sites for N-linked glycans; the GNS protein contains nine. The products of ephemerovirus α1, α2, β and γ genes have not been identified. The α1 gene product appears to be a viroporin but the functions of other products have not been established.
Antigenic properties
Ephemeroviruses cross-react strongly in CF or indirect immunofluorescence tests and may show low level cross-reactions by indirect immunofluorescence with viruses of the genus Lyssavirus. However, sequence comparisons with other rhabdoviruses indicate that in evolutionary terms the ephemeroviruses are closer to vesiculoviruses than to members of other genera in the family. There is only one known BEFV serotype worldwide, with virus isolates from different regions (Australia, Asia, Africa and the Middle-East) displaying significant cross-neutralization. There is a low level of cross-neutralization between BEFV and BRMV; there is no cross-neutralization between ARV and other ephemeroviruses. The BEFV G protein contains four distinct neutralization sites. The BEFV G protein purified from virions or expressed from recombinant vaccinia virus protects cattle from experimental infection. The GNS glycoprotein does not induce neutralizing antibodies and is not protective.
Biological properties
Bovine ephemeral fever is an economically important disease of cattle and water buffalo in most tropical and sub-tropical regions of Africa, Australia, the Middle-East and Asia. BEFV infection causes a sudden onset of fever and other clinical signs including lameness, anorexia and ruminal stasis, followed by a sustained drop in milk production. Although the mortality rate is usually low (1–2%), it is highest in well-conditioned beef cattle and high-producing dairy cattle. The virus is transmitted by, and replicates in, hematophagous arthropods and has been isolated from both culicoids and mosquitoes. Other species in the genus are not recognized as animal pathogens, but are known to infect cattle and have been isolated from healthy sentinel cattle.
Species demarcation criteria in the genus
Species cross-react in complement-fixation and/or indirect immunofluorescence tests but exhibit low to no cross-neutralization. They exhibit similar but distinct genome organizations with the common feature of a non-structural glycoprotein (GNS) gene but variations in the number of accessory protein genes and the location of transcriptional control sequences. Different species may share up to 91% identity in N protein amino acid sequence.
List of species in the genus Ephemerovirus
| Adelaide River virus |
|
|
| Adelaide River virus | [L09206*, L09208*, U05987*, U10363*] | (ARV) |
| Berrimah virus |
|
|
| Berrimah virus |
| (BRMV) |
| Bovine ephemeral fever virus |
|
|
| Bovine ephemeral fever virus BB7721 | [AF234533] | (BEFV-BB7721) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Ephemerovirus but have not been approved as species
| Kimberley virus | [AY854637*] | (KIMV) |
| Kotonkan virus | [HM474855] | (KOTV) |
| Malakal virus |
| (MALV) |
| Obodhiang virus | [HM856902] | (OBOV) |
| Puchong virus |
| (PUCV) |
* Sequences do not comprise the complete genome.
Genus Novirhabdovirus
Type species Infectious hematopoietic necrosis virus
Distinguishing features
This genus comprises one of the two major subgroups of rhabdoviruses known to infect aquatic hosts. Members of the other subgroup include spring viremia of carp virus (SVCV), which is a member of the genus Vesiculovirus, and several other viruses that have not been placed taxonomically.
Novirhabdoviruses have five major structural proteins, designated L (150–225 kDa), G (63–80 kDa), N (38–47 kDa), P (22–26 kDa, formerly designated M1), and M (17–22 kDa, formerly designated M2). In addition to the structural proteins, novirhabdoviruses encode a small, sixth, non-virion protein designated NV (12–14 kDa), which is expressed at variable levels in infected cells but is not detectable in purified virions. The NV ORF is preserved in numerous diverse viruses and strains, but the NV protein sequences are significantly less conserved between viruses in different species than sequences of the other structural proteins, such that there is no significant amino acid sequence similarity between the NV proteins of IHNV and VHSV. The specific function of the NV protein is not yet defined but it is required for efficient virus replication. Results of studies with NV gene deletion mutants generated by reverse genetics are inconsistent in that the NV appears to be required for pathogenicity in IHNV and VHSV but not SHRV.
Virion properties
Morphology
Virions are bullet-shaped and measure 45–100 nm in diameter×100–430 nm in length. Surface projections are densely dispersed, distinctive spikes which cover the whole surface except for the quasi-planar end.
Physicochemical and physical properties
The replication temperature range and thermal inactivation temperatures for these viruses are typically lower than those of other rhabdoviruses, due to the aquatic poikilotherm nature of their hosts. Optimum virus replication temperatures range from 15–28 °C, depending roughly on the ambient water temperature in the geographic range of each virus.
Nucleic acid
The genome comprises a single molecule of negative sense. single stranded RNA of about 11 kb.
Genome organization and replication
The genomic RNA is approximately 11.1 kb, with six genes in the order 3′-N-P-M-G-NV-L-5′. The genome contains a leader region of approximately 60 nt preceding the transcription start of the N gene, and a trailer of about 100 nt following the transcription termination of the L gene. Genes begin with the conserved putative transcription start signal 3′-CCRWG (vRNA sense, most often 3′-CCGUG), and the signal 3′-UUGU is also found upstream of the translation initiation site in many genes. Transcription terminates at the signal 3′-UCURUC(U)7, and non-transcribed intergenic regions are single nucleotides, G or A (vRNA sense).
Biological properties
Novirhabdoviruses infect fish of numerous species. The natural host ranges of individual viruses vary in breadth, with the type member IHNV limited to salmonid fish, while VHSV infects hosts from a wide range of fish families as diverse as salmonids and herring. In nature and in artificial environments novirhabdoviruses can be transmitted horizontally, from fish to fish, by a waterborne route. Egg-associated transmission has also been clearly demonstrated by several cases in which the spread of virus to new geographic regions has occurred with transport of contaminated eggs. It is increasingly apparent that wild fish can serve as reservoirs of virus. The existence of invertebrate reservoirs or vectors of virus has been postulated but their importance is uncertain. Similarly, the potential for a carrier state in survivors of IHNV infections has been demonstrated, but the frequency and significance of this phenomenon is not well understood.
The geographic distribution of novirhabdoviruses is broad. IHNV is enzootic to western North America, but inadvertent transport of the virus with contaminated eggs and infected fish has resulted in spread and establishment of IHNV in western Europe, Korea, Taiwan, Japan, and mainland China. VHSV is enzootic to cultured rainbow trout in much of western Europe, but more recently several North American and Asian strains have been described, and an extensive VHSV reservoir in marine fish in the northern Atlantic and Pacific Oceans has been demonstrated. Hirame rhabdovirus (HIRRV) is at present only isolated in Japan and Korea, and SHRV occurs in southeast Asia.
Members of the genus Novirhabdovirus cause disease in cultured fish hosts, resulting in significant economic losses to aquaculture industries. Both IHNV and VHSV have been well documented as severe pathogens of cultured salmonids since the 1950s, often resulting in losses of 50–100%. Among free-ranging fish IHNV epizootics have been reported in wild salmonids, and VHSV epizootics have occurred in both marine and freshwater fish of diverse species. IHNV and VHSV both cause hemorrhagic diseases, with petechial hemorrhages evident both externally and internally. Major degenerative changes and necrosis in the kidneys and hematopoietic tissue are evident upon histopathological examination, and are believed to be the actual cause of mortality.
Species demarcation criteria in the genus
Species within the genus have been distinguished serologically on the basis of cross-neutralization with polyclonal rabbit antisera. In general, strains within a species are neutralized by a single polyclonal antiserum. Thus, IHNV and HIRRV each comprise single serotypes, and VHSV has one major serotype with a small number of associated strains. Viruses from different species do not show cross-neutralization, but in some cases there is a low level of cross-reaction with specific proteins in western blot analyses. Nucleotide sequence data are available for most genes of these viruses, and will undoubtedly contribute to the distinction of viral species in the future. For strains within a virus species the nucleotide sequence divergence values range up to a maximum of 8% for IHNV G and NV genes, and 18% for the G genes of European and North American VHSV. N protein aa sequence identity between IHNV and VHSV is approximately 34%.
List of species in the genus Novirhabdovirus
| Hirame rhabdovirus |
|
|
| Hirame rhabdovirus CA9703 | [AF104985] | (HIRRV-CA9703) |
| Infectious hematopoietic necrosis virus |
|
|
| Infectious hematopoietic necrosis virus WRAC | [L40883] | (IHNV-WRAC) |
| Snakehead virus |
|
|
| Snakehead virus | [AF147498] | (SHRV) |
| Viral hemorrhagic septicemia virus |
|
|
| Egtved virus |
|
|
| Viral hemorrhagic septicemia virus Fil3 | [Y18263] | (VHSV-Fil3) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Novirhabdovirus but have not been approved as species
| Eel virus B12 |
| (EEV-B12) |
| Eel virus C26 |
| (EEV-C26) |
Plant-adapted rhabdovirus genera, Cytorhabdovirus and Nucleorhabdovirus
Distinguishing features
Two genera of plant rhabdoviruses have been established on the basis of the sites of virus replication and morphogenesis (Figure 5) (cytoplasm: Cytorhabdovirus; nucleus: Nucleorhabdovirus). Moreover, genus classification based on sequence diversity has thus far correlated 100% with classification by intracellular virus maturation. The interrelationships of the different plant viruses within or between the two genera or with the >50 putative plant rhabdoviruses (identified based on their unique particle morphology compared to other plant viruses) have largely yet to be established at the genetic level. There is no significant sequence similarity (>50%) between analogous genes of the species analyzed to date. A wide variety of plants are susceptible to rhabdoviruses although each virus usually has a restricted host range. Plant rhabdoviruses are transmitted by leafhoppers, plant-hoppers or aphids. Some viruses are also transmitted during vegetative propagation, and some can also be transmitted mechanically from infected sap. In all carefully examined cases, viruses have been shown to replicate in cells of the insect vector as well as in the plant host.
Genus Cytorhabdovirus
Type species Lettuce necrotic yellows virus
Distinguishing features
Cytorhabdoviruses replicate in the cytoplasm of infected cells in association with masses of thread-like structures (viroplasms). Virions bud in association with the endoplasmic reticulum (ER) and accumulate in ER-derived vesicles. A nuclear phase has been suggested but not proven in the replication of some cytorhabdoviruses (e.g. lettuce necrotic yellows virus (LNYV). Evidence of the nuclear involvement in the replication of others is lacking (e.g. barley yellow striate mosaic virus, BYSMV). Endogenous transcriptase activity is readily detectable in cytorhabdovirus preparations where this has been investigated.
Virion properties
Morphology
Enveloped, bacilliform virions, 60–75 nm in diameter and 200–350 nm long.
Physicochemical and physical properties
Buoyant density of 1.19–1.20 g cm−3 in sucrose or potassium tartrate. The lipid envelope is derived from the cytoplasmic membranes of plant or insect host cells.
Nucleic acid
Monopartite, negative sense, single stranded RNA genome, 12.8–14.5 kb in length. Six to ten mRNAs, one for each of the encoded proteins identified in infected plants.
Genome organization and replication
The genome of LNYV is about 12.8 kb and the genome organization is similar to that of SYNV (see Nucleorhabdovirus). Preceded by a non-coding 84 nt leader sequence, the gene order is 3′-N-P-4b-M-G-L-5′. N represents the nucleoprotein, P, M, G and L are the putative phospho-, matrix- and glycoproteins and RNA polymerase, respectively. Protein 4b has been predicted to represent a movement protein. The intergenic regions contain highly conserved consensus sequences. The 5′-non-coding trailer sequence of 187 nt has extensive complementarity to the 3′-leader. The genome of northern cereal mosaic virus (NCMV) is about 13.2 kb with a gene order similar to that of LNYV, except for the presence of three additional small genes of unknown function between P and M and an additional gene between G and L. The genome organization of lettuce yellow mottle virus (LYMoV) (12.9 kb) and strawberry crinkle virus (SCV) (14.5 kb) are similar to that of LNYV, except for the presence in SCV of one additional gene between G and L.
Species demarcation criteria in the genus
In the genus Cytorhabdovirus, species are primarily differentiated by plant host range and vector specificity of the virus. Nucleic acid hybridization has been used to provide confirmation of species and serological criteria have enabled verification of common viruses that infect different hosts. However, no virus strains have been defined unambiguously using serology. The complete nucleotide sequence is available for only four viruses in the genus, LNYV, LYMoV, NCMV and SCV. Thus, this criterion is not presently sufficient for discrimination of species. Hybridization using cloned probes and conserved L gene polymerase motif sequences has been used to differentiate viruses within the genus and to identify some strains. These analyses should be emphasized in future studies.
List of species in the genus Cytorhabdovirus
| Barley yellow striate mosaic virus |
|
|
|
| Barley yellow striate mosaic virus Zanjan-1 | {planthopper} | [FJ665628*] | (BYSMV-Z1) |
| Maize sterile stunt virus | {planthopper} |
| (MSSV) |
| Wheat chlorotic streak virus | {planthopper} |
| (WCSV) |
| Broccoli necrotic yellows virus |
|
|
|
| Broccoli necrotic yellows virus | {aphid} |
| (BNYV) |
| Festuca leaf streak virus |
|
|
|
| Festuca leaf streak virus |
|
| (FLSV) |
| Lettuce necrotic yellows virus |
|
|
|
| Lettuce necrotic yellows virus 318 | {aphid} | [AJ867584] | (LNYV-318) |
| Lettuce yellow mottle virus |
|
|
|
| Lettuce yellow mottle virus - France |
| [EF687738] | (LYMoV-FRA) |
| Northern cereal mosaic virus |
|
|
|
| Northern cereal mosaic virus - Hebei | {planthopper} | [GU985153] | (NCMV-Hb) |
| Sonchus virus |
|
|
|
| Sonchus virus |
|
| (SonV) |
| Strawberry crinkle virus |
|
|
|
| Strawberry crinkle virus - UK | {aphid} | [AY005146*, AY250986*] | (SCV-UK) |
| Wheat American striate mosaic virus |
|
|
|
| Wheat American striate mosaic virus | {leafhopper} |
| (WASMV) |
| Oat striate mosaic virus | {leafhopper} |
| (OSMV) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ], natural vector species { } and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Cytorhabdovirus but have not been approved as species
| Wheat rosette stunt virus | {planthopper} | [AF059602-04*, AF059677*, AF064784*] | (WRSV) |
| Soybean blotchy mosaic virus | {leafhopper} | [EU877231*] | (SbBMV) |
| Ivy vein banding virus |
| [GQ249162*, GQ249163*] | (IVBV) |
* Sequences do not comprise the complete genome.
Genus Nucleorhabdovirus
Type species Potato yellow dwarf virus
Distinguishing features
Nucleorhabdoviruses replicate in the nuclei of plant cells, which become greatly enlarged and develop large granular nuclear inclusions that are thought to be sites of virus replication. In situ hybridization analyses have shown that the viral genomic and antigenomic RNAs are highly expressed in subnuclear foci and immunofluorescence studies have shown that the N, P and L nucleocapsid also accumulate in subnuclear foci. Viral proteins are synthesized from discrete poly-adenylated mRNAs and reporter gene analyses have shown that they accumulate in subnuclear foci. Virus morphogenesis occurs at the inner nuclear membrane, and enveloped virus particles accumulate in perinuclear spaces. In protoplasts treated with tunicamycin, morphogenesis is interrupted and nucleocapsids accumulate in the nucleoplasm.
Virion properties
Morphology
Enveloped, bacilliform virions, 45–100 nm in diameter and 130–300 nm long.
Physicochemical and physical properties
Virus particles sediment at 800–1000S in sucrose gradients and the buoyant density of virions is 1.18 g cm−3 in isopycnic sucrose gradients.
Nucleic acid
Monopartite, negative sense, single stranded RNA genome, 12–14 kb in length. Six to seven mRNAs, one for each of the encoded proteins identified in infected plants.
Genome organization and replication
The genome of potato yellow dwarf virus (PYDV) is about 12.9 kb long and encodes seven ORFs in the order 3′-N-X-P-Y-M-G-L-5′, which likely encode the nucleocapsid (N), phospho- (P), movement (Y), matrix (M), glyco- (G) and RNA-dependent RNA polymerase (L) proteins, respectively. The function of X has not been determined. The ORFs are flanked by a 3′ leader RNA of 149 nt and a 5′ trailer RNA of 97 nt, and are separated by conserved intergenic “gene junction” regions which are similar in length and have sequence relatedness with those of other rhabdoviruses. M protein is able to induce the intranuclear accumulation of the inner nuclear membrane in the absence of any other viral protein. Protein interaction studies in live plants have identified binary interactions between N:N, N:P, M:M, M:Y, M:G, G:G and Y:Y.
The genome of sonchus yellow net virus (SYNV)(ca. 13.7 kb), 3′-N-P-sc4-M-G-L-5′, lacks an X protein. The Y equivalent, named sc4, is believed to be involved in cell-to-cell movement. The 144 nt 3′ leader sequence is transcribed to produce a polyadenylated leader RNA, which localizes in the cytoplasm. The 5′ trailer RNA is 160 nt long with extensive terminal complementarity with the leader sequence. The N and P proteins contain nuclear localization sequences (NLS) and are independently imported into the nucleus, where they associate and move to a sub-nuclear location. A distinct nuclear polymerase complex composed of N, P and L is present in the nuclei of infected cells.
The genome of rice yellow stunt virus (RYSV) is 14 kb with a gene order similar to that of SYNV, except for the presence of an additional gene between G and L which encodes a virion-associated protein. The genomes of maize mosaic virus (MMV) and taro vein chlorosis virus (TaVCV) are about 12 kb with a gene order similar to that of SYNV. The genome of maize fine streak virus (MFSV) is 13.8 kb with a gene order similar to that of PYDV, with an ORF of unknown function between P and M.
Species demarcation criteria in the genus
Species are primarily differentiated by plant host range and vector specificity of the virus. Nucleic acid hybridization has been used to provide confirmation of identification and serological criteria have enabled verification of common viruses that infect different hosts. However, no virus strains have been defined unambiguously using serology. The complete nucleotide sequences are available for viruses in six species of the genus (MFSV, MMV, PYDV, RYSV, SYNV, TaVCV) and partial sequences have been determined for eggplant mottled dwarf virus. With additional sequences becoming available in the near future, nucleotide sequences, RT-PCR-based assays and fluorescent viral protein localization may become useful tools for species demarcation. Hybridization using cloned probes has been used to verify viruses within the genus and these and other molecular analyses should be emphasized in future studies.
List of species in the genus Nucleorhabdovirus
| Datura yellow vein virus |
|
|
|
| Datura yellow vein virus |
|
| (DYVV) |
| Eggplant mottled dwarf virus |
|
|
|
| Eggplant mottled dwarf virus-Egg | {leafhopper} | [AM922319*, AM922322*] | (EMDV-Egg) |
| Pittosporum vein yellowing virus |
|
| (PVYV) |
| Tomato vein yellowing virus |
|
| (TVYV) |
| Pelargonium vein clearing virus |
|
| (PVCV) |
| Maize fine streak virus |
|
|
|
| Maize fine streak virus - USA | {leafhopper} | [AY618417] | (MFSV-US) |
| Maize mosaic virus |
|
|
|
| Maize mosaic virus - USA | {planthopper} | [AY618418*] | (MMV-US) |
| Potato yellow dwarf virus |
|
|
|
| Potato yellow dwarf virus - SYDV | {leafhopper} | [GU734660] | (PYDV-SYDV) |
| Rice yellow stunt virus |
|
|
|
| Rice yellow stunt virus - China | {leafhopper} | [AB011257] | (RYSV-China) |
| Rice transitory yellowing virus - Ishigaki | {leafhopper} | [AB516283] | (RTYV-Japan) |
| Sonchus yellow net virus |
|
|
|
| Sonchus yellow net virus - USA | {aphid} | [L32603] | (SYNV-US) |
| Sowthistle yellow vein virus |
|
|
|
| Sowthistle yellow vein virus - USA | {aphid} |
| (SYVV-US) |
| Taro vein chlorosis virus |
|
|
|
| Taro vein chlorosis virus - Fiji |
| [AY674964] | (TaVCV-FJ) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ], natural vector species { } and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Nucleorhabdovirus but have not been approved as species
| Cereal chlorotic mottle virus | {leafhopper} |
| (CCMoV) |
| Cynodon rhabdovirus |
| [EU650683*] | (CRV) |
| Maize Iranian mosaic virus | {planthopper} | [DQ186554] | (MIMV) |
| Sorghum stunt mosaic virus | {leafhopper} |
| (SSMV) |
* Sequences do not comprise the complete genome.
List of unassigned species in the family Rhabdoviridae
| Flanders virus |
|
|
|
| Flanders virus - USA | {Culex spp. (mosquitoes)} | [AF523194-9] | (FLAV-US) |
| Ngaingan virus |
|
|
|
| Ngaingan virus MRM14556 | {Culicoides brevitarsis} | [FJ715959] | (NGAV- MRM14556) |
| Sigma virus |
|
|
|
| Sigma virus HAP23 | {Drosophila melanogaster} | [GQ375258] | (SIGMAV-HAP23) |
| Sigma virus AP30 | {Drosophila melanogaster} | [NC_013135] | (SIGMAV-AP30) |
| Tupaia virus |
|
|
|
| Tupaia virus - Thailand | {Tupaia belangeri} | [AY840978] | (TUPV-TH) |
| Wongabel virus |
|
|
|
| Wongabel virus CS264 | {Culicoides austropalpalis} | [EF612701] | (WONV-CS264) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ], natural vector species { } and assigned abbreviations ( ) are also listed.
Sigma virus (SIGMAV) naturally infects fruit flies (Drosophila spp.) and is transmitted vertically through the germinal cells. Infected flies exposed to carbon dioxide are irreversibly paralyzed. Based on its genome organization, phylogenetic placement and pathobiology SIGMAV appears to be unique and not a member of any previously described genus. Virions are spiked and enveloped bullet-shaped particles of about 75×140–200 nm appear to be exclusively cytoplasmic and contain a helical nucleocapsid. The genome contains six genes arranged in the order 3′-N-P-X-M-G-L-5′. There is an additional gene between P and M genes, like viruses in the genera Cytorhabdovirus and Nucleorhabdovirus; the encoded putative protein is of unknown function but contains conserved domains found in reverse transcriptases. Another unusual feature is that M and G mRNAs overlap by 33 nucleotides.
Flanders virus (FLAV) is associated with mosquitoes and birds in North America. Serological studies demonstrated that FLAV isolates from different states of the USA were closely related to each other, but different from Hart Park virus (HPV). Furthermore, FLAV was found mostly in eastern North America, whereas HPV was found mostly in western North America. Some overlap is found at the geographic extremes but, for the most part, these viruses have been found where their mosquito hosts occur, Culiseta melanura in the east (FLAV), Culex tarsalis in the west (HPV). FLAV genome is about 13 kb with a gene order of 3′-N-(tc)-P-(tc)-pseudogene1-(tc)-19K-(tc)-pseudogene2-(tt)-M-G-(tt)-L-5′. The unique features include the additional 19K gene, surrounded by two pseudogenes, about 500 nucleotides each, situated between the P and M genes. According to the N and L gene sequences, FLAV demonstrates limited relatedness to WONV and NGAV viruses,which circulate in Australia, and this cluster is distantly related to ephemeroviruses (Figure 6).
Tupaia virus (TUPV) was isolated from spontaneously degenerating hepatocellular carcinoma cells from a tree shrew (Tupaia belangeri), imported from Thailand and kept in captivity for about 6 years. The host range of the virus in vitro appears to be restricted to tupaia cells. The genome is about 11.5 kb with a gene order 3′-N-P/C-M-SH-G-L-5′. The genome contains a unique small hydrophobic (SH) transcription unit between M and G genes, and the corresponding transcript was found in the infected cells. The combined transmembrane topology and signal peptide prediction algorithms identified the SH as a type I transmembrane protein with a signal peptide, a small extracellular domain, a transmembrane region, and a cytoplasmic tail. Furthermore, the overlapping ORF in the TUPV P mRNA has the potential to code for a 221-amino-acid C protein. Based on the N gene phylogeny, TUPV is mostly related to unassigned viruses African Kolongo virus and Sandjimba virus, both isolated from birds (Figure 6). In other analyses, where limited fragments of the L gene were compared, TUPV demonstrated relatedness to Humpty Doo virus, isolated from gnats in Australia. Therefore, viruses related to TUPV may be distributed quite broadly.
List of other related animal viruses which may be members of the family Rhabdoviridae but have not been approved as species
| Almpiwar virus | {Ablepharus boutonii (lizard)} | [AY854645*] | (ALMV) |
| Aruac virus | {Trichoprosopon theobaldi (mosquito)} |
| (ARUV) |
| Bahia Grande virus |
|
| (BGV) |
| Bangoran virus | {Culex perfuscus} |
| (BGNV) |
| Barur virus | {Rattus rattus wroughtoni} |
| (BARV) |
| Bimbo virus | {Euplectes afra (bird)} |
| (BBOV) |
| Bivens Arm virus | {Culicoides insignis (midge)} |
| (BAV) |
| Blue crab virus |
|
| (BCV) |
| Chaco virus | {Ameiva ameiva (lizard)} |
| (CHOV) |
| Charleville virus | {Phlebotomus spp. (sandfly)} | [AY854644*] | (CHVV) |
| Coastal Plains virus | {Bos taurus} |
| (CPV) |
| Connecticut virus | {Ixodes tentatus (tick)} |
| (CNTV) |
| Cuiaba virus | {Bufo marinus (toad)} |
| (CUIV) |
| Curionopolis virus | {Culicoides spp.} |
| (CURV) |
| DakArK 7292 virus | {Amblyomma variegatum (tick)} |
| (DAKV-7292) |
| Durham virus | {Fulica americana (bird)} |
| (DURV) |
| Entamoeba virus |
|
| (ENTV) |
| Farmington virus | {bird} |
| (FARV) |
| Fukuoka virus | {Culicoides punctatus} | [AY854651*] | (FUKAV) |
| Garba virus | {Corythornis cristata (bird)} |
| (GARV) |
| Gossas virus | {Tadarita sp. (bat)} |
| (GOSV) |
| Harlingen virus | {Culex spp . (mosquitoes)} |
| (HARV) |
| Hart Park virus | {Culex tarsalis} |
| (HPV) |
| Humpty Doo virus | {Lasiohelea sp.(midge)} | [AY854643*] | (HDOOV) |
| Iriri virus | {Lutzomyia spp. (sandfly)} |
| (IRIV) |
| Inhangapi virus | {Lutzomyia flaviscutellata} |
|
|
| Itacaiunas virus | {Culicoides spp. (midge)} |
| (ITAV) |
| Joinjakaka virus | {Mixed Culicine mosquitoes} |
| (JOIV) |
| Kamese virus | {Culex annulioris} |
| (KAMV) |
| Kannamangalam virus | {Corvus splendens (crow)} |
| (KANV) |
| Kern Canyon virus | {Myotis yumanensis (bat)} | [DQ457101*] | (KCV) |
| Keuraliba virus | {Tatera kempi (gerbil)} |
| (KEUV) |
| Kolongo virus | {Euplecies afra (bird)} | [DQ457100*] | (KOLV) |
| Koolpinyah virus | {bovine} |
| (KOOLV) |
| Landjia virus | {Riparia paludicola (bird)} |
| (LJAV) |
| Le Dantec virus | {human} | [AY854650*] | (LDV) |
| Manitoba virus | {Culex tarsalis} |
| (MNTBV) |
| Marco virus | {Ameiva ameiva} |
| (MCOV) |
| Morreton virus | {Lutzomyia sp.} |
| (MRTV) |
| Mosqueiro virus | {Culex portesi} |
| (MQOV) |
| Mossuril virus | {Culex sitiens} |
| (MOSV) |
| Mount Elgon bat virus | {Rhinolophus sp. (bat)} | [DQ457103*] | (MEBV) |
| Moussa virus | {Culex decens} | [FJ985748] | (MOUV) |
| Muir Springs virus | {Aedes sp. (mosquitoes)} |
| (MSV) |
| Nasoule virus | {Andropadus virens (bird)} |
| (NASV) |
| Navarro virus | {Cathartes aura (bird)} |
| (NAVV) |
| New Minto virus | {Haemaphysalis leporispalustris (tick)} |
| (NMV) |
| Nkolbisson virus | {Eretmapodites leucopus (mosquito)} |
| (NKOV) |
| Oak-Vale virus | {Culex spp.} | [GQ294474*] | (OVRV) |
| Oita virus | {Rhinolophus cornutus (bat)} | [AB116386*] | (OITAV) |
| Ouango virus | {Sitagra melanocephala (bird)} |
| (OUAV) |
| Parry Creek virus | {Culex annulirostris} | [AY854647*] | (PCRV) |
| Reed Ranch virus | {Culex sp.} |
| (RRV) |
| Rio Grande cichlid virus |
|
| (RGRCV) |
| Rochambeau virus | {Coquillettidia albicosta} | [DQ457104*] | (RBUV) |
| Sandjimba virus | {Acrocephalus schoenobaenus (bird)} | [DQ457102*] | (SJAV) |
| Sawgrass virus | {Dermacentor variabilis} |
| (SAWV) |
| Sea trout rhabdovirus |
| [AF434992*] | (STRV) |
| Sena Madureira virus | {Ameiva ameiva} |
| (SMV) |
| Siniperca chuatsi rhabdovirus |
| [DQ399789] | (SCRV) |
| Sripur virus | {Sergentomyia spp. (sandflies)} |
| (SRIV) |
| Sweetwater Branch virus |
|
| (SWBV) |
| Tibrogargan virus | {Culicoides brevitarsis} | [GQ294472] | (TIBV) |
| Timbo virus | {Ameiva ameiva} |
| (TIMV) |
| Xiburema virus | {Sabethes intermedius} |
| (XIBV) |
| Yata virus | {Mansonia uniformis} |
| (YATAV) |
Virus names are in roman script. Sequence accession numbers [ ], natural vector species { } and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome. For most of the listed viruses, no biochemical characterization has been reported. Their listing here is largely based on the distinctive morphology of their virions.
List of other related plant viruses which may be members of the family Rhabdoviridae but have not been approved as species
| Atropa belladonna virus |
| (AtBV) |
| Beet leaf curl virus | {lacebug} | (BLCV) |
| Callistephus chinensis chlorosis virus |
| (CCCV) |
| Carnation bacilliform virus |
| (CBV) |
| Carrot latent virus | {aphid} | (CtLV) |
| Cassava symptomless virus |
| (CsSLV) |
| Chrysanthemum frutescens virus |
| (CFV) |
| Chrysanthemum vein chlorosis virus |
| (CVCV) |
| Clover enation virus |
| (ClEV) |
| Colocasia bobone disease virus | {planthopper} | (CBDV) |
| Coriander feathery red vein virus | {aphid} | (CFRVV) |
| Cow parsnip mosaic virus |
| (CPaMV) |
| Cynara virus |
| (CraV) |
| Dendrobium leaf streak virus |
| (DLSV) |
| Digitaria striate virus | {planthopper} | (DiSV) |
| Euonymus fasciation virus |
| (EFV) |
| Finger millet mosaic virus | {planthopper} | (FMMV) |
| Gerbera symptomless virus |
| (GeSLV) |
| Gomphrena virus |
| (GoV) |
| Holcus lanatus yellowing virus |
| (HLYV) |
| Iris germanica leaf stripe virus |
| (IGLSV) |
| Ivy vein clearing virus |
| (IVCV) |
| Laelia red leafspot virus |
| (LRLV) |
| Launea arborescens stunt virus |
| (LArSV) |
| Lemon scented thyme leaf chlorosis virus |
| (LSTCV) |
| Lolium ryegrass virus |
| (LoRV) |
| Lotus stem necrosis |
| (LoSNV) |
| Lotus streak virus | {aphid} | (LoSV) |
| Lucerne enation virus | {aphid} | (LEV) |
| Lupin yellow vein virus |
| (LYVV) |
| Maize streak dwarf virus |
| (MSDV) |
| Malva silvestris virus |
| (MaSV) |
| Melilotus latent virus |
| (MeLV) |
| Melon variegation virus |
| (MVV) |
| Parsley virus |
| (PaV) |
| Phalaenopsis chlorotic spot virus |
| (PhCSV) |
| Pigeon pea proliferation virus | {leafhopper} | (PPPV) |
| Pineapple chlorotic leaf streak virus |
| (PCLSV) |
| Pisum virus |
| (PisV) |
| Plantain mottle virus |
| (PIMV) |
| Ranunculus repens symptomless virus |
| (RaRSV) |
| Raphanus virus |
| (RaV) |
| Raspberry vein chlorosis virus | {aphid} | (RVCV) |
| Red clover mosaic virus |
| (RCIMV) |
| Saintpaulia leaf necrosis virus |
| (SLNV) |
| Sambucus vein clearing virus |
| (SVCV) |
| Sarracenia purpurea virus |
| (SPV) |
| Soursop yellow blotch virus |
| (SYBV) |
| Triticum aestivum chlorotic spot virus |
| (TACSV |
| Vigna sinensis mosaic virus |
| (VSMV) |
| Winter wheat Russian mosaic virus | {planthopper} | (WWMV) |
| Zea mays virus |
| (ZMV) |
Virus names are in roman script. Natural vector species { } and assigned abbreviations ( ) are also listed.
There are many putative plant rhabdoviruses. Many of these agents have not been characterized beyond electron microscopic observations in infected plants and occasionally in their vectors. Hence, their listing here relies on morphological criteria. Some have been transmitted experimentally by mechanical means and by their vectors.
Phylogenetic relationships within the family
Molecular phylogenies determined by using N, G, or L protein sequences suggest a monophyletic origin and support the assignment of the six established genera and the species within these genera. For P and G proteins, relatively low sequence identities across the family prevent the construction of a universal phylogeny. N and L protein sequences are most highly conserved (see Figure 6). Both phylogenetic analyses indicate that vesiculoviruses and ephemeroviruses are the most closely related of the established genera and together with some currently non-classified viruses may be considered as members of a phylogenetic supergroup named “Dimarhabdovirus” (dipteran-mammal associated rhabdovirus).
Similarity with other taxa
Rhabdoviruses share several features with viruses of the families Filoviridae, Paramyxoviridae and Bornaviridae in the order Mononegavirales. Features they have in common include the non-segmented negative sense, single strand, non-infectious RNA genome, the helical nucleocapsid with the genome template intimately associated with the nucleoprotein (RNase resistant), the initiation of primary transcription by a virion-associated RdRp, similar gene order and single 3′ promoter with short terminal untranscribed regions and intergenic regions. The virions are large enveloped structures with a prominent fringe of spikes. They generally replicate in the cytoplasm (except nucleorhabdoviruses). They mature by budding, predominantly from the plasma membrane with the exception of RABV which buds occasionally from internal membranes and plant rhabdoviruses of the genus Nucleorhabdovirus which bud from the inner nuclear membrane. They transcribe discrete unprocessed messenger RNAs for which they ensure 5′-capping and 3′-polyadenylation. Polymerase amino acid sequence similarities with those of plant-infecting viruses outside the Mononegavirales include viruses in the genera Ophiovirus and Varicosavirus and the unclassified orchid fleck virus.
Derivation of names
Cyto: from Greek kytos, “cell”.
Ephemero: from Greek ephemeros, “short-lived”.
Lyssa: from Greek lyssa, “rage, fury, canine madness”.
Novi: from non-virion protein.
Nucleo: from Latin nux, nucis, “nut”.
Rhabdo: from Greek rhabdos, “rod”.
Vesiculo: from Latin vesicula, diminutive of vesica, “bladder, blister”.
Further reading
Books and journals
Ammar et al., 2009 E-D. Ammar, C.-W. Tsai, A.E. Whitfield, M.G. Redinbaugh, S.A. Hogenhout, Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 54 (2009) 447–468.
Bandyopadhyay et al., 2010 A. Bandyopadhyay, K. Kopperud, G. Anderson, K. Martin, M. Goodin, An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology. 402 (2010) 61–71.
Bootland and Leong, 1999 L.M. Bootland, J.C. Leong, Infectious hematopoietic necrosis virus. In: . CAB International, New York, NY199957–121.
Bourhy et al., 2008 H. Bourhy, A.J. Gubala, R.P. Weir, D.B. Boyle, Animal rhabdoviruses. In: . Elsevier, Oxford2008111–121.
Hoffmann et al., 2005 B. Hoffmann, M. Beer, H. Schütze, T.C. Mettenleiter, Fish rhabdoviruses: molecular epidemiology and evolution. Curr. Top. Microbiol. Immunol. 292 (2005) 81–117.
Jackson and Wunner, 2007 A.C. Jackson, W.H. Wunner, . In: A.C. Jackson, W.H. Wunner, . Academic Press/Elsevier, London2007.
Jackson et al., 2005 A.O. Jackson, R.G. Dietzgen, M.M. Goodin, J.N. Bragg, M. Deng, Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43 (2005) 623–660.
Smail, 1999 D.A. Smail, Viral haemorrhagic septicaemia. In: . CAB International, New York, NY1999123–146.
Stone et al., 2003 D.M. Stone, W. Ahne, A.M. Sheppard, C.T-Y. Tsin-yee Liu, G.R. Taylor, K.D. Denham, P.F. Dixon, K. Way, Nucleotide sequence analysis of the glycoprotein gene of putative spring viraemia of carp viruses and pike fry rhabdovirus isolates reveals four distinct piscine vesiculovirus genogroups. Dis. Aquat. Org. 53 (2003) 203–210.
Walker, 2008 P.J. Walker, Bovine ephemeral fever virus. In: . Elsevier, Oxford2008354–362.
Websites
The Centers for Disease Control and Prevention, Rabies program: http://www.cdc.gov/rabies
IHNV epidemiology and genetic typing database: http://gis.nacse.org/ihnv
North American VHSV epidemiology and genetic typing database: http://gis.nacse.org/vhsv
Fish Pathogens EU database of genetic data on fish viruses IHNV and VHSV: http://www.fishpathogens.eu/vhsv/index.php
Contributed by
Dietzgen, R.G., Calisher, C.H., Kurath, G., Kuzmin, I.V., Rodriguez, L.L., Stone, D.M., Tesh, R.B., Tordo, N., Walker, P.J., Wetzel, T. and Whitfield, A.E.
Figures
Figure 1 (Left) Diagram illustrating a rhabdovirus virion and the nucleocapsid structure (courtesy of P. Le Mercier); (right) negative contrast electron micrograph of virions of an isolate of vesicular stomatitis Indiana virus. The bar represents 100 nm.
(Courtesy of P. Perrin.)
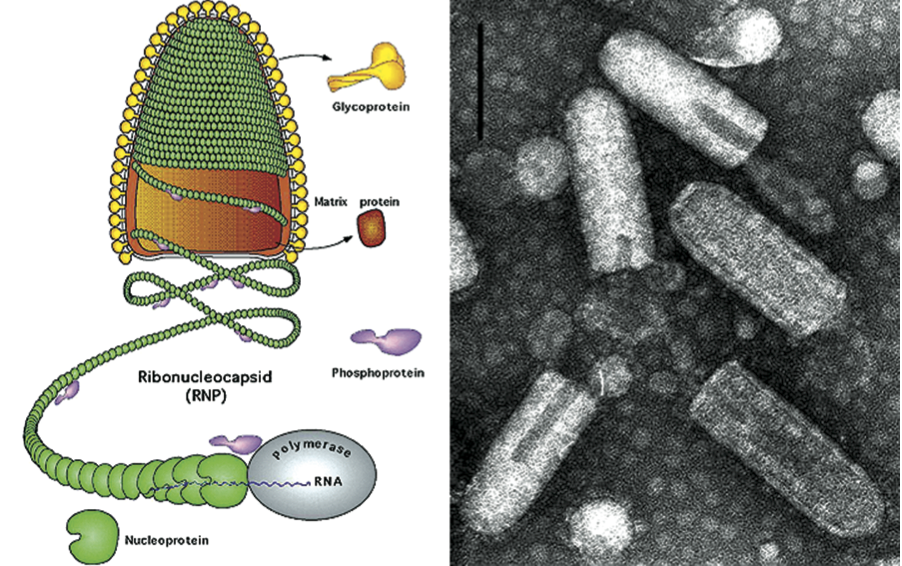
Figure 2 Comparison of genome organization of representative members from all genera of the family Rhabdoviridae. A modular organization into three blocks has been conserved: the 3 block #1 encodes N/P proteins required in large amounts; the central block #2 encodes the membrane M/G proteins; the 5 block #3 encodes the polymerase L required in limited amounts. Between blocks, viruses have inserted typical genes adapted to their particular biology, for example the movement protein (3, 4b) in plant rhabdoviruses.
(Courtesy of N. Tordo.)
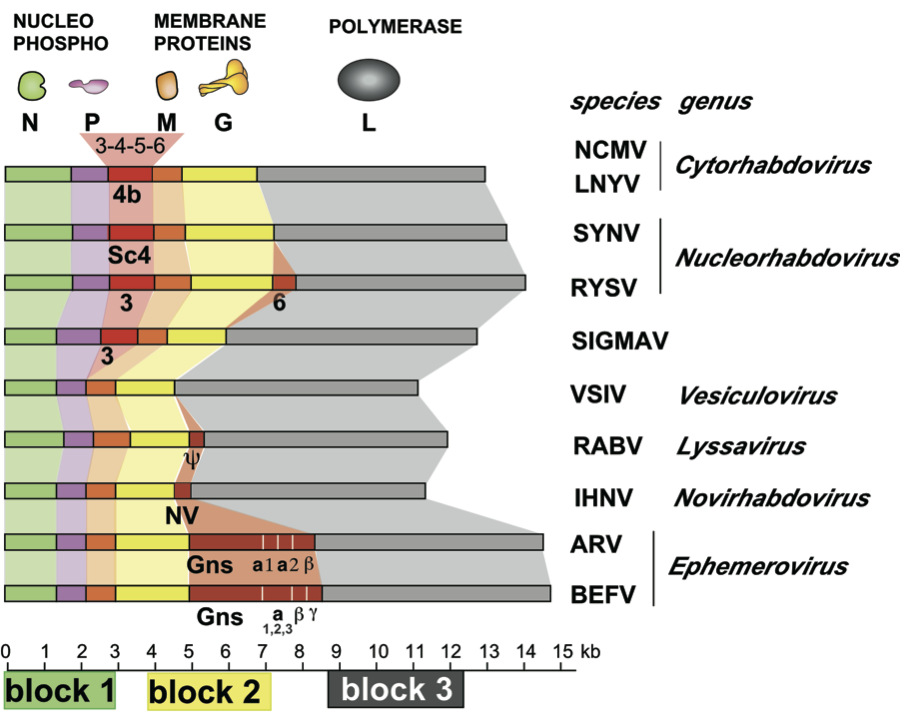
Figure 3 Panel (1) shows the genome organization of vesicular stomatitis Indiana virus (VSIV) and the process of consecutive transcription of leader RNA and monocistronic mRNAs. Panel (2) illustrates the replication of the negative sense genome via a positive sense antigenome intermediate. The switch from transcription to replication appears to be regulated by the N protein.
(Courtesy of P. Le Mercier.)
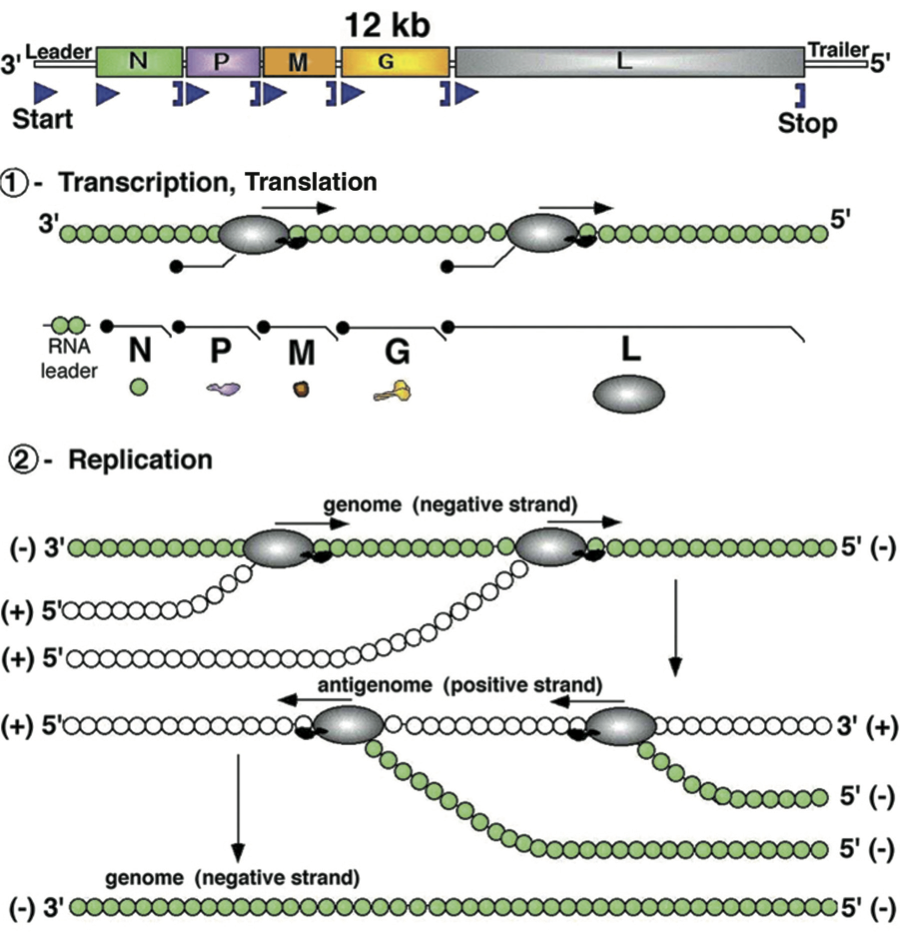
Figure 4 Architecture of the vesicular stomatitis virus virion. Montage model of the tip and cryoEM map of the trunk. N is green, M is blue, and the inner (2) and outer (1) leaflets of the membrane are violet and pink. (Inset) illustration of the base region of the VSV virion. The X marks the absence of a turn of M helix below the lowest turn of the N helix.
(From Ge, P., Tsao, J., Schein, S., Green, T. J., Luo, M. and Zhou, Z. H. (2010). Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science, 327, 68993; with permission.)
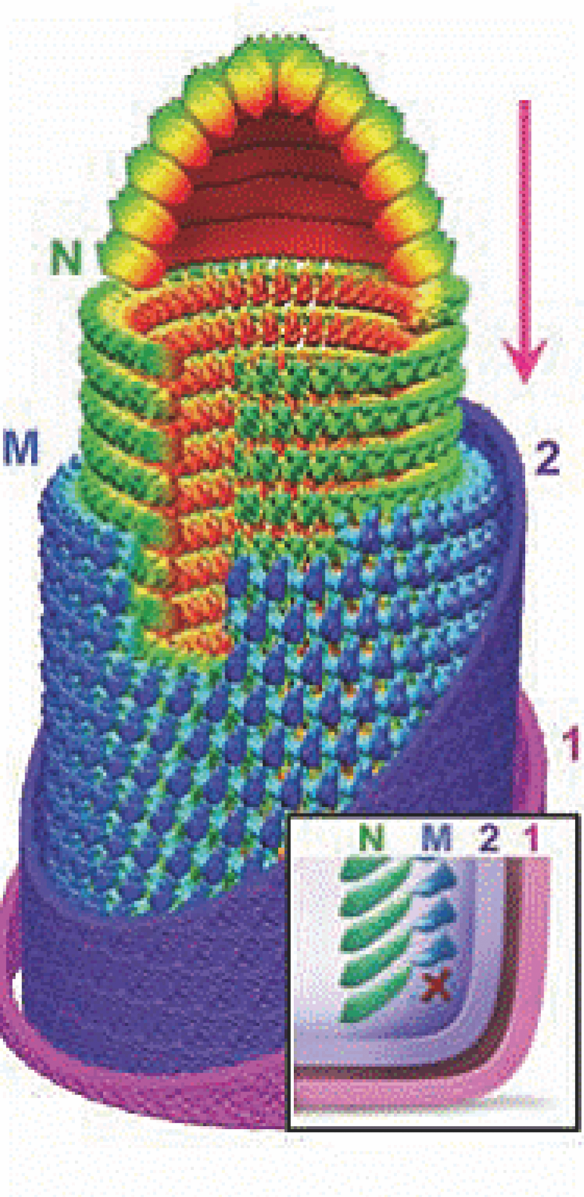
Figure 5 Contrasts between the replication cycles of cytorhabdoviruses and nucleorhabdoviruses. Most rhabdoviruses gain entry into host cells during insect vector feeding. Uncoating is believed to take place on ER membranes, followed by release of the nucleocapsid core into the cytoplasm. At this point, the replication cycles of the two genera diverge. In the case of the cytorhabdoviruses, the newly released cores become transcriptionally active and associate with the endoplasmic reticulum to establish viroplasms that function in transcription of viral mRNAs (vmRNAs) and replication of genomic and antigenomic viral RNAs. Following translation of the vmRNAs, viral proteins involved in replication accumulate in the viroplasm. Viral glycoproteins are targeted to cytoplasmic membranes or, possibly, the outer nuclear envelope (ONE). Maturation of cytorhabdoviruses takes place via matrix protein-mediated condensation of cores at sites of G protein accumulation in the endoplasmic reticulum. In the case of the nucleorhabdoviruses, released cores are transported into the nucleus through nuclear pore complexes (NPC). Following transcription and export, vmRNAs are translated and viral proteins are imported into the nucleus, where they participate in replication and formation of large viroplasms. During intermediate stages of infection of plant rhabdoviruses, movement of infectious units from cell to cell occurs. Nucleocapsids most likely are the transported form, and these interact with viral encoded movement proteins that participate in number of activities, including nucleocapsid binding, transport through the NPC to the plasmodesmata, and modifications to the plasmodesmatal size exclusion limits. Morphogenesis occurs near the end of active transcription and replication and involves interactions with the M protein to coil the viral nucleocapsids and form associations with membrane-associated G protein. In the cytorhabdoviruses, electron microscopic observations suggest that budding occurs into proliferated ER associated with the viroplasms. Currently, at least two models can be proposed for morphogenesis of nucleorhabdovirus virions. In recent data outlined in the text, the inner nuclear envelope (INE) proliferates due to redistribution of cytoplasmic membranes and invaginates to form intranuclear spherules, into which viral budding occurs. In the classical model, virus budding occurs through intact INE resulting in an expansion of the outer nuclear envelope. In both models, mature virions accumulate in the perinuclear spaces of infected cells where they may be reacquired during subsequent insect feeding.
(Reproduced from Jackson et al. (2005); with permission from Annual Reviews.)
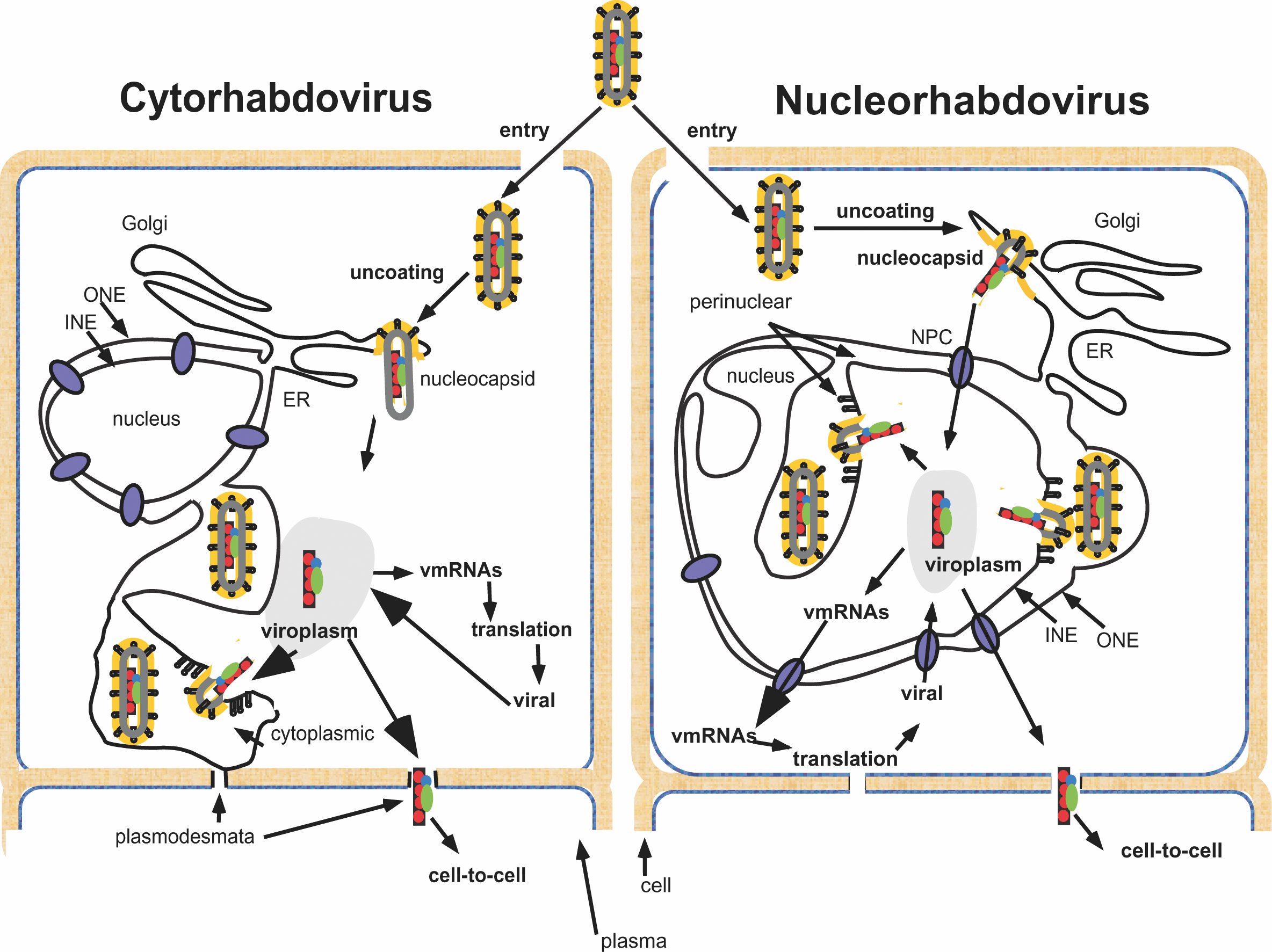
Figure 6 Phylogenetic tree of viruses in the Rhabdoviridae based on alignment of nucleoprotein gene sequences. The tree was generated by the neighbor-joining method using 1000 bootstrap replicates.