Order: Mononegavirales
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
The virions are large enveloped structures with a prominent fringe of peplomers, 5–10 nm long and spaced 7–10 nm apart, in all except the members of the family Bornaviridae. The morphologies are variable and individual particles frequently exhibit pleomorphism, but in general distinguish the families: 90 nm diameter spherical particles with a 50 nm diameter electron-dense core and without peplomers in the family Bornaviridae; simple, branched, U-shaped, 6-shaped or circular filaments of uniform diameter (about 80 nm) extending up to 14,000 nm are characteristic of viruses classified in the family Filoviridae, although purified virions are bacilliform and of uniform length (e.g. 790 nm in the case of Marburg virus); filamentous, pleomorphic or spherical structures of variable diameter are characteristic of viruses belonging to the family Paramyxoviridae; and regular bullet-shaped or bacilliform particles are characteristic of the member viruses of the family Rhabdoviridae. The ribonucleoprotein core has a diameter of 13–20 nm, which in viruses belonging to the families Filoviridae and Rhabdoviridae is organized into a helical nucleocapsid of about 50 nm in diameter. Differences in helical pitch of the ribonucleoprotein core distinguish the Paramyxovirinae from the Pneumovirinae.
Physicochemical and physical properties
Virion Mr is 300–1000×106. S20,W is 550–1045S (plant rhabdoviruses have larger S20,W values). Virion buoyant density in CsCl 1.18–1.22 g cm−3. Virus infectivity is rapidly inactivated by heat treatment at 56 °C, or following UV- or X-irradiation, or exposure to lipid solvents.
Nucleic acid
Virions contain one molecule of linear, non-infectious, negative sense, ssRNA, 8.9–19 kb in size, Mr of 3–5×106 which comprises about 0.5 to 2.0% of the particle weight. The viral RNA lacks a capped 5’ terminus, or a covalently associated protein. The 3’ terminus of viral RNA lacks a poly(A) tract. The 5’- and 3’-terminal regions exhibit significant inverse complementarity, and there are conserved motives in the terminal regions of all four families. Full-length positive sense (anti-genomic) RNAs are found in infected cells. The genome comprises a linear sequence of genes, with limited overlaps in some viruses, and with short terminal non-coding regions. The virus genes are expressed as individual transcription units, each bounded by short transcription start and termination sequences. The transcription start and termination sequences show considerable similarities within the families with conserved motifs present in all families. The non-transcribed intergenic regions range from two to several hundred nucleotides. The predominant pattern is that each individual virus mRNA encodes a single protein but exceptions are seen; genetic information may be encoded in all three reading frames in the P genes of respiroviruses and morbilliviruses and the M2 genes of the Pneumovirinae encode two proteins from different reading frames. Splicing of some mRNA and overlapping start/stop signals are characteristic of bornaviruses. In the subfamily Paramyxovirinae of the family Paramyxoviridae, but not the subfamily Pneumovirinae, the number of nucleotides in the genome is divisible by six (“the rule of six”), presumably reflecting a nucleocapsid structural constraint.
Proteins
There are a limited number of proteins in relation to the large particle size. The 5–7 structural proteins comprise envelope glycoprotein(s), a matrix protein, a major RNA-binding protein, other nucleocapsid-associated protein(s), plus a large molecular weight polymerase protein, and in some viruses several non-structural proteins which may be phosphorylated. The matrix protein is non-glycosylated in all except the bornaviruses. The matrix protein of Borna disease virus is N-glycosylated and expressed on the surface of virions. Enzymatic activities associated with the virions may include transcriptase, polyadenylate transferase, mRNA transferase and neuraminidase.
Lipids
Virions are composed of about 15–25% lipids, their composition reflecting that of the host cell membrane from which the virions bud. Generally, phospholipids represent about 55–60%, and sterols and glycolipids about 35–40% of the total lipids. Glycoproteins may have a covalently associated fatty acid proximal to the lipid envelope.
Carbohydrates
Virions are composed of about 3% carbohydrate by weight. The carbohydrates are present as N- and O-linked glycan chains on surface proteins and on glycolipids. When made in mammalian cells the oligosaccharide chains are generally of the complex type, in insect cells they are of the non-complex types.
Genome organization and replication
In the families Filoviridae, Paramyxoviridae and Rhabdoviridae, the site of multiplication is the cytoplasm, with the exception of viruses classified in the genus Nucleorhabdovirus. The 3’ end (leader) region of the genomes contains the promoter element which directs the virus RdRp to initiate transcription. Discrete mRNAs are transcribed by sequential interrupted synthesis. Transcription is polar, with a gradient of attenuation. The order of the genes on the genome encoding the structural proteins is conserved, though individual members may carry additional genes located between the structural genes (Table 1). The presence of these additional genes may affect the relative levels of the mRNAs from the structural genes. The mRNAs are capped and polyadenylated, with polyadenylation occurring as a result of iterative transcription from a short poly U tract at the end of each gene. Generally, genes do not overlap, the exceptions being the M2 and L genes of pneumoviruses, the VP30 and VP24 of the Marburgviruses and the VP35/VP40, GP/VP30 and VP24/L of ebolaviruses. The P genes of Paramyxovirinae encode multiple proteins. In members of the respirovirus, morbillivirus and Henipavirus genera, the C proteins arise from the use of an alternative reading frame accessed by a standard AUG initiation codon, and additional C-related proteins in the respiroviruses by the use of non-AUG start codons. Members of the avulavirus and rubulavirus genera do not encode a C protein. In all members of the subfamily Paramyxovirinae, except human parainfluenzavirus type 1, a proportion of the P mRNA produced by the virus is altered by an editing process in which one or more additional guanine residues, not represented by cytosine residues in the template genome, are inserted at a specific site in the mRNA during transcription. This process results in translation of the edited mRNA utilizing a different reading frame following the position of the inserted base(s) to direct the synthesis of the additional V and W proteins. In the avulaviruses the faithfully transcribed mRNA encodes the V protein and the P protein is produced from an edited mRNA. A non-templated insertion event occurs during transcription of the glycoprotein gene of ebolaviruses generating both membranes-inserted and secreted forms of the glycoprotein. The M2 mRNA of the members of the Pneumovirinae sub-family encode a second protein from an additional ORF accessed by a coupled translation process dependent on translation of the upstream ORF. Replication occurs by synthesis of a complete positive sense anti-genomic RNA. Genomic and anti-genomic RNAs are present as nucleocapsids. In the family Bornaviridae, the site of multiplication is the nucleus. Transcription of bornavirus genomes is complex, with splicing of mRNA and overlapping stop/start signals. The mRNAs of bornaviruses are capped, but synthesis is not inhibited by alpha-amanitin, suggesting that a cap-snatching mechanism is not involved. In the filoviruses, paramyxoviruses and rhabdoviruses, maturation of the independently assembled helical nucleocapsid occurs by budding through host membranes with investment by a host-derived lipid envelope containing transmembrane proteins. The process of assembly and maturation of bornaviruses is not known at present.
Table 1 A diagrammatic representation of the 3’ to 5’ arrangement of the transcriptional units in the genomes of viruses classified in the four families (Bornaviridae, Filoviridae, Paramyxoviridae and Rhabdoviridae) comprising the order Mononegavirales
|
Family/Subfamily |
Genus |
Virus |
3’←Gene order→5’ |
||||||||||||||
|
Bornaviridae |
Bornavirus |
BDV |
le |
|
|
N |
X/P |
|
M |
|
|
|
G |
|
|
L |
tr |
|
Rhabdoviridae |
Vesiculovirus |
VSV |
le |
|
|
N |
P |
|
M |
|
|
|
G |
|
|
L |
tr |
|
|
Lyssavirus |
RabV |
le |
|
|
N |
P |
|
M |
|
|
|
G |
Ps |
|
L |
tr |
|
|
Cytorhabdovirus |
LNYV |
le |
|
|
N |
P |
4b |
M |
|
|
|
G |
|
|
L |
tr |
|
|
Nucleorhabdovirus |
SYNV |
le |
|
|
N |
P |
Sc4 |
M |
|
|
|
G |
|
|
L |
tr |
|
|
Novirhabdovirus |
IHNV |
le |
|
|
N |
P |
|
M |
|
|
|
G |
NV |
|
L |
tr |
|
|
Ephemerovirus |
BEFV |
le |
|
|
N |
P |
|
M |
|
|
|
G |
Gns |
|
L |
tr |
|
Filoviridae |
Ebolavirus |
ZeboV |
le |
|
|
N |
P |
|
M1 |
|
|
|
GP/SP |
|
M2 |
L |
tr |
|
|
Marburgvirus |
MarV |
le |
|
|
N |
P |
|
M1 |
|
|
|
G |
|
M2 |
L |
tr |
|
Paramyxoviridae |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Paramyxovirinae |
Avulavirus |
NDV |
le |
|
|
N |
P/V |
|
M |
F |
|
|
H |
|
|
L |
tr |
|
|
Henipavirus |
HeV |
le |
|
|
N |
P/C/V |
|
M |
F |
|
|
H |
|
|
L |
tr |
|
|
Morbillivirus |
MeV |
le |
|
|
N |
P/C/V |
|
M |
F |
|
|
H |
|
|
L |
tr |
|
|
Respirovirus |
SeV |
le |
|
|
N |
P/C/V |
|
M |
F |
|
|
HN |
|
|
L |
tr |
|
|
Rubulavirus |
MuV |
le |
|
|
N |
V/P |
|
M |
F |
|
SH |
HN |
|
|
L |
tr |
|
Pneumovirinae |
Metapneumovirus |
AMPV |
le |
|
|
N |
P |
|
M1 |
F |
M2 |
SH |
G |
|
|
L |
tr |
|
|
Pneumovirus |
HRSV |
le |
NS1 |
NS2 |
N |
P |
|
M1 |
SH |
G |
F |
M2 |
|
|
L |
tr |
|
Unassigned viruses |
|
JV |
le |
|
|
N |
P/C/V |
|
M |
F |
|
SH |
G |
|
|
L |
tr |
|
|
|
BeV |
le |
|
|
N |
P/C/V |
|
M |
F |
|
SH |
G |
|
|
L |
tr |
|
|
|
FDLV |
le |
|
|
N |
P/V |
|
M |
F |
|
|
HN |
|
|
L |
tr |
Genes encoding polypeptides of presumed homologous function are aligned vertically. The genome organizations of currently unclassified viruses which encode additional proteins are also shown.Abbreviations: viruses: AMPV, avian metapneumovirus; BDV, Borna disease virus; BeV, Beilong virus; BEFV, bovine ephemeral fever virus; FDLV, Fer de Lance virus; HeV, Hendra virus; HRSV, human respiratory syncytial virus; IHNV, infectious hemopoetic necriosis virus; JV, J virus; LNYV, lettuce necrotic yellows virus; MarV, Lake Victoria marburgvirus; MeV, measles virus; MuV, mumps virus; NDV, Newcastle disease virus; RabV, rabies virus; SeV, Sendai virus; SYNV, sonchus yellow net virus, VSV, vesicular stomatitis Indiana virus; ZeboV, Zaire ebolavirus.Transcriptional units; le, non-coding leader region; NS, non-structural protein gene; N, nucleoprotein gene; U, gene of unknown function; P, phosphoprotein gene; V and C, dispensible non-structural protein genes; sc4 and 4b, genes of unknown function; M and M1, non-glycosylated matrix protein gene; (M) glycosylated matrix protein gene; F, fusion protein gene; SH, small hydrophobic protein gene; TM, transmembrane protein gene; G (or H or HN), glycosylated (or hemagglutinin or hemagglutinin/neuraminidase) attachment protein gene; M2, non-glycosylated (BDV excepted) envelope protein gene; Ps, pseudogene; NV, non-virion protein gene; Gns, presumptive duplicated G sequence; GP/SP, the glycoprotein gene of ebolavirus produces two products, the major secreted form from the unaltered mRNA and the structural form produced by transcriptional insertion of a single base in the mRNA; L, large (polymerase) protein gene; tr, non-coding trailer region.
Antigenic properties
The major neutralizing epitopes against which antibodies are directed lie within the membrane glycoproteins. Virus serotypes are defined by the surface antigens. Filoviruses are an exception in that they are poorly neutralized in vitro. In bornaviruses, antibodies to both the glycosylated matrix protein, which may function as an attachment protein, and the gp94 envelope protein neutralize infectivity.
Biological properties
The host ranges vary from restricted to unrestricted. Filoviruses have only been isolated from primates and swine. Paramyxoviruses occur only in vertebrates and no vectors are known. Rhabdoviruses infect invertebrates, vertebrates and plants. Some rhabdoviruses multiply in both invertebrates and vertebrates, some in invertebrates and plants, but none in all three. In human hosts the pathogenic potential tends to be characteristic of the family: i.e. hemorrhagic fever (Filoviridae); respiratory and neurological diseases (Paramyxoviridae); mild febrile to fatal neurological diseases (Rhabdoviridae). Bornaviruses have been isolated from horses, cattle, sheep, rabbits, rats, cats, ostriches and man. The pathology associated with virus infection is variable. Infection of animals is associated with conditions ranging from behavioral disturbances to severe non-purulent encephalomyelitis. Cytopathology varies from none (bornaviruses and filoviruses) to rapidly lytic (rhabdoviruses and paramyxoviruses); syncytium formation is common in paramyxoviruses.
Phylogenetic relationships within the order
Phylogenetic relationships between the families Bornaviridae, Filoviridae, Paramyxoviridae and Rhabdoviridae are illustrated in Figure 1.
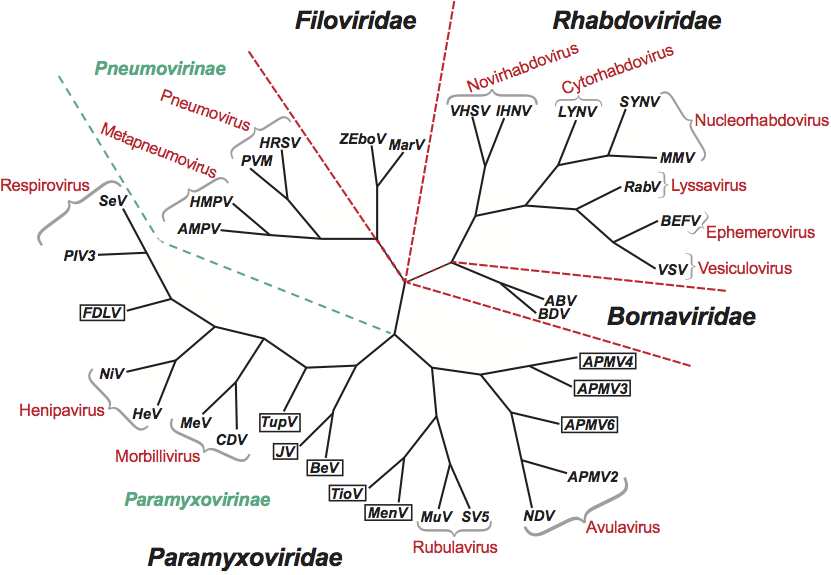
Figure 1 Unrooted phylogenetic tree of members of the order Mononegavirales. The tree was constructed using the CLUSTALX and PHYLIP programs with the sequences of the conserved domain III of the polymerase proteins (Poch et al., 1989, 1990). Nine paramyxoviruses, not yet assigned to genera, are included (boxed): avian parainfluenza virus types 3 (APMV3), avian parainfluenza virus types 4 (APMV4), avian parainfluenza virus types 6 (APMV6), Beilong virus (BeV), Fer de Lance virus (FDLV), J virus (JV), Menangle virus (MenV), Tioman virus (TioV) and Tupaia paramyxovirus (TupV). Abbreviations: ABV, avian bornavirus; AMPV, avian metapneumovirus; APMV2, avian parainfluenza virus types 2; BDV, Borna disease virus; BEFV, bovine ephemeral fever virus; CDV, canine distemper virus; HeV, hendra virus; HMPV, human metapneumovirus; HRSV, human respiratory syncytial virus; IHNV, infectious hemorraghic necrosis virus; LNYV, lettuce necrotic yellows virus; MarV, Marburg virus; MeV, measles virus; MMV, maize mosaic virus; MuV - mumps virus; NDV, Newcastle disease virus; NiV, Nipah virus; PIV3, parainfluenza virus type 3; PVM, pneumonia virus of mice; RabV, rabies virus; SeV, Sendai virus; SV5, simian virus 5; SYNV, Sonchus yellow net virus; VSV, vesicular stomatitis Indiana virus; VHSV, viral hemorrhangic septicemia virus; ZeboV, Zaire ebolavirus.
Derivation of names
Borna: from Borna, a town in Saxony.
Cyto: from Greek kytos, “cell”.
Ebola: from the river Ebola, in Sudan and Zaire.
Ephemero: from Greek ephemeros, “short-lived”.
Filo: from Latin filo, “thread-like”.
Lyssa: from Greek lyssa, “rage, fury, canine madness”.
Marburg: from the city of Marburg, in Germany.
Meta: from Greek meta, “after”.
Mono: from Greek monos, “single”.
Morbilli: from Latin morbillus, diminutive of morbus, “disease”.
Nega: from negative sense RNA.
Novi: non virion protein gene, characteristic of the genus.
Nucleo: from Latin Nux, nucis “nut”.
Paramyxo: from Greek para, “by the side of”, and myxa, “mucus”.
Pneumo: from Greek pneuma, “breath”.
Respiro: from Latin respirare, “to breathe”.
Rhabdo: from Greek rhabdos, “rod”.
Rubula: from Latin ruber, “red”; Rubula inflans, old name for mumps.
Vesiculo: from Latin vesicula, diminutive of vesica, “bladder”, “blister”.
Virales: from Latin virales, “viruses”.
Further reading
De la Torre, 1994 J.C. De la Torre, Molecular biology of Borna disease virus: Prototype of a new group of animal viruses. J. Virol. 68 (1994) 7669–7675.
Mulberger et al., 1992 E. Mulberger, A. Sanchez, A. Randolf, C. Will, M.P. Kiley, H. Klenk, H. Feldmann, The nucleotide sequence of the L gene of Marburg virus, a filovirus: homologies with paramyxoviruses and rhabdoviruses. Virology. 187 (1992) 534–547.
Poch et al., 1990 O. Poch, B.M. Blumberg, L. Bougueleret, N. Tordo, Sequence Comparisons of five polymerase (L-proteins) of unsegmented negative-strand RNA viruses – theoretical assignment of functional domains. J. Gen. Virol. 71 (1990) 1153–1162.
Poch et al., 1989 O. Poch, I. Sauvaget, M. delaRue, N. Tordo, Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8 (1989) 3867–3874.
Pringle, 1997 C.R. Pringle, The order Mononegavirales – current status. Arch. Virol. 142 (1997) 2321–2326.
Pringle and Easton, 1997 C.R. Pringle, A.J. Easton, Monopartite negative strand RNA genomes. Semin. Virol. 8 (1997) 49–57.
Schneemann et al., 1995 E. Schneemann, P.A. Schneider, R.A. Lamb, W.I. Lipkind, The remarkable coding strategy of Borna disease virus: a new member of the non-segmented negative strand RNA viruses. Virology. 210 (1995) 1–9.
Tomonaga et al., 2002 K. Tomonaga, T. Kobayashi, K. Ikuta, Molecular and cellular biology of Borna disease virus infection. Microb. Infect. 4 (2002) 491–500.
Tordo et al., 2005 N. Tordo, P-E. Ceccaldi, Y. Gaudin, A.I. Wandeler, Rhabdoviruses: rabies. In: . Hodder Arnold, London20051102–1136.
Wang et al., 2000 L.-F. Wang, M. Yu, E. Hansson, L.I. Pritchard, B. Shiell, W.P. Michalski, B. Eaton, The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J. Virol. 74 (2000) 9972–9979.
Contributed by
Easton, A.J. and Pringle, C.R.
