Family: Bornaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Bornavirus
Type species Borna disease virus
Virion properties
Morphology
Electron microscopy studies of negatively stained infectious particles of an isolate of Borna disease virus (BDV) have shown that virions have a spherical morphology with a diameter of 90±10 nm containing an internal electron-dense core (50–60 nm) (Figure 1).
Physicochemical and physical properties
Virion Mr and the S20,w are not known. Partially purified BDV infectious particles have a buoyant density in CsCl of 1.16–1.22 g cm−3, in sucrose of 1.22 g cm−3, in renografin of 1.13 g cm−3. Virus infectivity is rapidly lost by heat treatment at 56 °C. Virions are relatively stable at 37 °C, and only minimal infectivity loss is observed after 24 hrs incubation at 37 °C in the presence of serum. Virions are inactivated below pH 5.0, as well as by treatment with organic solvents, detergents, and exposure to UV radiation. Infectivity is completely and rapidly destroyed by chlorine-containing disinfectants or formaldehyde treatment.
Nucleic acid
The genome consists of a single molecule of a linear, negative sense ssRNA about 8.9 kb in size and Mr of about 3×106). The RNA genome is not polyadenylated. Extracistronic sequences are found at the 3′ (leader) and 5′ (trailer) ends of the BDV genome. BDV 3′-terminal genomic sequences have a high A+U content with a U/A ratio of about 2:1. The ends of the BDV genome RNA exhibit partial inverted complementarity. Full-length plus-strand (antigenomic) RNAs are present in infected cells and in viral ribonucleoproteins. Defective RNAs have not been identified in BDV-infected cells and tissues. BDV can be classified into two subtypes based on the complete genome sequences of several BDV strains. With the unique exception of strain No/98, all isolates of BDV to date (independent of year, species and area isolation) have approximately 95% homology at the nt level (subtype 1), while BDV strain No/98 shows only 85–86% nt sequence identity compared to other BDV strains (subtype 2). The nt changes are distributed fairly evenly over the entire genome of BDV. However, all strains predict the same BDV genomic organization and differ by only one nt in absolute genome size.
Proteins
Six major ORFs are found in the BDV genome sequence (Figure 2). These ORFs code for polypeptides with predicted size of 40 kDa (p40), 24 kDa (p24), 10 kDa (p10), 16 kDa (p16), 56 kDa (p56) and 180 kDa (p180), respectively. Based on their positions in the viral genome and abundance in infected cells and virion particles, together with their biochemical and sequence features, p40, p24 and p16 BDV polypeptides correspond to the viral nucleoprotein (N), the phosphoprotein (P) transcriptional activator, and matrix (M) proteins, respectively, found in other negative sense ssRNA viruses. Two isoforms of the BDV N (p39 and p38) are found in BDV-infected cells. These two forms of the viral N appear to be encoded by two different mRNA species. Differential usage of two in-frame initiation codons present in the BDV p40 gene may also contribute to the production of BDV p39/38. BDV p39 contains both a nuclear localization signal (NLS) and a nuclear export signal (NES), whereas p38 harbors only the NES. BDV p24 is an acidic polypeptide (predicted I.P. of 4.8), that has a high Ser-Thr content (16%), with phosphorylation at serine residues which is mediated by both protein kinase C and casein kinase II. These features are consistent with those of the phosphoprotein (P) transcriptional activator found in other negative sense ssRNA viruses. BDV p24 contains a bipartite NLS in the sequence. In addition to P, a 16 kDa polypeptide (P′) is also translated from the second in-frame AUG codon in the P ORF. An additional ORF, p10, encodes a polypeptide of 10 kDa present in BDV-infected cells. BDV X starts within the same mRNA transcription unit, 46 nt upstream from p24 and overlaps, in a different frame, with the 71 N-terminal aa of p24. Recent study indicates that BDV X harbors a NLS in the N-terminus of the sequence.
Consistent with other negative sense ssRNA viruses, BDV p16, the putative BDV M protein, is a non-glycosylated M protein, associated at the inner surface of the viral membrane. BDV ORF4 (p56) overlaps, in a different frame, with the C-terminus of ORF p16, and is capable of encoding a 503 aa polypeptide with a predicted size of 56 kDa. Based on its sequence features, BDV p56 is the counterpart of the virus surface glycoproteins (G) found in other negative sense ssRNA viruses. The p56 gene directs the synthesis of three glycosylated polypeptides of about 84 or 94 kDa (GP-84/94, G), 43 kDa (GP-43, GP-C) and 45 to 55 kDa (GP-N). G corresponds to the full length of the p56 gene, whereas GP-C and GP-N represent the C-terminal subunit and the N-terminal subunit of ORF p56, respectively. Both GP-C and GP-N are associated with BDV infectious particles. Antibodies to p56 have neutralizing activity, suggesting that BDV p56 gene products play an important role in the early steps of BDV infection. BDV ORF5 (p180) is capable of encoding a polypetide with a predicted size of 180 kDa, whose deduced aa sequence displays strong homology to other negative sense ssRNA virus polymerases, members of the L protein family. An additional ORF predicted in mRNA species generated via RNA splicing would encode a variant BDV L with a predicted size of 190 kDa (BVp190). BDVp190 corresponds to BDVp180 with 153 aa added to its N-terminus. Recent evidence suggests that p190, rather than p180, is the active BDV L. BDV L contains the NLS in the sequence.
Lipids
Not known.
Carbohydrates
Only N-glycans, mannose-rich type and partially hybrid types.
Genome organization and replication
The negative sense BDV RNA genome codes for at least six ORFs in the order 3′-N-P/X-M-G-L-5′. The genomic polarity has a very limited coding capability, and none of its predicted ORFs has a favorable translational start signal; further they are not flanked by putative transcription start and termination/polyadenylation signals. Therefore, it seems unlikely that BDV uses an ambisense coding strategy. BDV has the property, unique among known negative sense ssRNA animal viruses, of a nuclear site for genome transcription and replication. Full-length genome complementary RNA molecules (antigenomes) act as templates for new viral genome RNA synthesis. Genome and anti-genome RNA molecules are neither capped nor polyadenylated. These RNAs exist as nucleocapsids in the nucleus of infected cells. It is unknown whether RNA species corresponding to the leader RNA are transcribed in BDV-infected cells.
BDV cell entry occurs by receptor-mediated endocytosis. The virus G protein has been implicated in entry. The identity of the BDV cellular receptor is unknown. In endosomes, low pH-dependent fusion occurs between viral and cellular membranes. This fusion event releases the BDV ribonucleoproteins (RNP) which are then transported to the cell nucleus where viral transcription and replication occur. Sequential and polar transcription results in decreasing molar quantity of BDV transcripts from the 3′- to the 5′-encoded cistrons. The viral mRNAs are polyadenylated, and their 5′ ends contain a blocking group, presumably a cap structure. Virus specific mRNA synthesis is not inhibited by α-amanitin, and sequences at the 5′ of the BDV mRNAs are homogeneous and genome-encoded. Thus, it is unlikely that transcription initiation of BDV mRNAs involves a cap-snatching mechanism similar to the one used by influenza viruses. Monocistronic viral mRNAs in BDV-infected cells are detected only for the N gene (Figure 2). The BDV G and L polymerase gene products are synthesized from downstream ORFs within polycistronic mRNAs. Mapping of BDV sgRNAs present in infected cells to the viral genome revealed that the BDV genome contains three transcription initiation sites (S signals), and four transcription termination/polyadenylation sites (E signals) (Figure 2). In addition, a putative E signal (E5) is found at nt 4776. The S signals contain a semi-conserved U-rich motif that is partially copied into the respective transcripts. A similar motif is not found within the S signals of previously described negative sense ssRNA viruses. BDV E signals consist of six or seven U residues preceded by a single A residue, resembling the E signal motif found in other negative sense ssRNA viruses. The BDV genome lacks the characteristic configuration of E signal/intergenic (IG) region/S signal, found at the gene boundaries of other negative sense ssRNA viruses. Instead, BDV transcription units and transcriptive signals frequently overlap (Figure 2). Two of the BDV primary transcripts are post-transcriptionally processed by the cellular RNA splicing machinery. Three introns (I, II and III) have been identified in the BDV genome. BDV introns I and II span nt 1932 to 2025 and 2410 to 3703, respectively, in the BDV antigenomic sequence (Figure 2). Splicing of intron I places the aa in position 13 of M next to a stop codon, whereas splicing of intron II, and I+II, results in a mRNA containing a predicted ORF that corresponds to the first 58 aa of G fused to a new C-terminus of 20 aa. RNA species resulting from splicing of intron II, and I+II, predicts also an additional ORF that would encode a variant BDV L protein with 153 aa added to the N-terminus. Intron III is generated by alternative 3′ splice site choice and spans nt 2410 to 4559 in the BDV antigenome (Figure 2). Splicing of introns II and III is regulated by the utilization of an alternative E signal (E5) and a putative cis-acting exon splicing suppressor signal located within the L gene. Transcripts lacking Intron III have the capacity to encode two new proteins with predicted size of 8.4 kDa (p8.4) and 165 kDa (p165). Whether these new predicted BDV polypeptides are synthesized in infected cells is unknown. BDV strain No/98 lacks the alternative 3′ splice site and thus cannot generate transcripts lacking intron III. RNA splicing can also modulate the efficiency of termination-reinitiation of translation and leaky scanning mechanisms, thus contributing to the regulation of the expression of BDV M, G and L gene products.
BDV-infected cells exhibit a heterogeneous pattern of viral antigen expression. BDV N, P and X polypeptides are expressed both in the nucleus and cytoplasm. N and P are the viral antigens expressed at higher levels, and they are expressed by the majority of the cells within an infected population. In contrast, only 1 to 10% of the infected cells express detectable levels of BDV G. Expression of full-length BDV G (GP-84/94) is restricted to the ER and nuclear envelope. The subcellular distribution of the BDV M is cytosolic and associated with cellular membranes. G is post-translationally modified by N-glycosylation. BDV G undergoes post-translational cleavage by the cellular protease furin, with the resulting GP-N and GP-C reaching the cell surface. Cleavage of G likely occurs in the trans-Golgi compartment. G, GP-N and GP-C are partially Endo H-sensitive and PNGase F-sensitive. The newly exposed N-terminus of GP-C is highly hydrophobic, and BDV-infected cells form extensive syncytia upon low-pH treatment. These findings suggest that GP-C is involved in pH-dependent fusion after internalization of BDV by receptor-mediated endocytosis. GP-N is most likely responsible for receptor binding.
The mechanisms involved in nucleocytoplasmic transport of viral RNP through the nuclear pore complex remain largely unknown. However, recent studies suggest that the nuclear import activity of BDV is mediated by the NLS-containing viral antigens, such as N, P, X and L, that form complexes in infected cells. In contrast, a nuclear export activity is found only in N protein. The NES of BDV N contains the canonical leucine-rich motif, and the nuclear export activity of the protein is mediated through the chromosome region maintenance protein (CRM1) pathway.
The assembly process and site of virus maturation have not been identified and budding of BDV particles from infected cells has been documented only from the surface of BDV-infected MDCK cells after treatment with n-butyrate. BDV RNP accumulate in the nucleus and, as with other negative sense ssRNA viruses, they are also infectious on the basis of an ability to direct synthesis of BDV macromolecules, as well as the production of BDV cell-associated infectivity upon transfection of BDV-susceptible cells. Thin sections of BDV-infected cells revealed the presence of intracytoplasmic virus-like particles with morphological characteristics similar to those described for partially purified cell-free BDV infectious particles. These particles showed no association with cisternae of the endoplasmic reticulum, the Golgi complex, or other intracytoplasmic membranes.
As for many other member of the order Mononegavirales, a reverse genetics system has recently been established for BDV.
Antigenic properties
BDV possess a number of distinct antigenic determinants. The so-called soluble antigen (s-antigen) obtained from the supernatant after ultracentrifugation of ultrasonicated BDV-infected brain tissue, contains the viral N, P and M proteins. Serum antibodies from BDV-infected animals frequently recognize all the components of the s-antigen, but rarely recognize the viral G products. BDV field isolates from the same or different animal species, as well as viruses recovered from experimental infections with different histories of passages exhibit strong serological cross-reactivity. There is only one recognized serotype of BDV, but monoclonal antibodies have revealed minor antigenic differences among BDV isolates. Complement independent IgG-specific neutralizing antibodies have been documented in experimentally infected animals. Titers of neutralizing antibodies are usually very low and dependent on the infected host species. BDV G protein has been implicated in virus neutralization.
Biological properties
Horses and sheep have been regarded as the main natural hosts of BDV. In these species BDV can cause a fatal neurologic disease, Borna disease (BD). Evidence indicates that the natural host range of BDV is wider than originally thought. Naturally occurring BDV infections have been documented in cattle, rabbits and cats. In addition, sporadic cases of natural infection with BDV have been reported in several other species, including donkeys, mules and llamas. Moreover, experimental infections have revealed a remarkable wide host range for BDV, from birds to rodents and non-human primates. BDV-induced neurobehavioral abnormalities in animals are reminiscent of some human neuropsychiatric disorders. Serological data and molecular epidemiological studies indicate that BDV can infect humans, and is possibly associated with certain neuropsychiatric disorders.
BDV is thought to be transmitted through salival, nasal, or conjunctival secretions. Infection may therefore result from direct contact with these secretions. Intranasal infection is the most likely route of natural infection, allowing BDV access to the central nervous system (CNS) by intraaxonal migration through the olfactory nerve. Cases of Borna disease (BD) are more frequent in some years than others and tend to occur in spring and early summer, suggesting arthropods as a potential vector. BDV has not been isolated from insects, but ticks have been implicated in the transmission of an infectious encephalomyelitis similar to BD affecting ruminants in the Middle East.
Asymptomatic naturally infected animals of different species have been documented in Europe, North America, Africa and Asia, suggesting that the prevalence and geographic distribution of BDV may have been underestimated. However, a definite natural reservoir of BDV has not been identified. Phenotypic differences have been described among different BDV field isolates, and among viruses with different histories of passages in animals and cultured cells. Despite its wide host range and phenotypic variation, molecular epidemiological data have shown a remarkable sequence conservation of BDV, not only within the same host species but also amongst sequences derived from different animal species.
BDV is highly neurotropic and has a non-cytolytic strategy of multiplication. BDV causes CNS disease in several non-human vertebrate species, which is characterized by neurobehavioral abnormalities that are often, but not always, associated with the presence of inflammatory cell infiltrates in the brain. BDV exhibits a variable period of incubation, from weeks to years, and diverse pathological manifestations that depend on the genetics, age and immune status of the host, as well as route of infection and viral determinants. Classic BD is caused by a T cell-dependent immune mechanism. Inflammatory cells are found forming perivascular cuffs and also within the brain parenchyma. Both CD4+ and CD8+ T-cells are present in the CNS cell infiltrates and contribute to the immune-mediated pathology associated with BD. BDV can also induce distinct deficiencies in emotional and cognitive functions that are associated with specific neurochemical disturbances in the absence of lymphoid infiltration. Heightened viral expression in limbic system structures, together with astrocytosis and neuronal structural alterations within the hippocampal formation are the main histopathological hallmarks of BDV infection.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Bornavirus
| Borna disease virus |
|
|
| Borna disease virus-V | [U04608] | (BDV-V) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Bornavirus but have not been approved as species
None reported.
Similarity with other taxa
BDV has a genomic organization similar to that of other negative sense ssRNA viruses. The size of the BDV genome (ca. 8.9 kb) is significantly smaller than those of the other known members of the order Mononegavirales: Rhabdoviridae (ca. 11–15 kb), Paramyxoviridae (ca. 15 kb) and Filoviridae (ca. 19 kb). BDV replication and transcription take place in the nucleus. This is a unique feature among known negative sense ssRNA animal viruses, but shared with the plant nucleorhabdoviruses. Expression of the BDV genome is regulated by an overlap of transcription units and transcriptive signals, an overlap of ORFs, readthrough of transcription termination signals and differential use of translational initiation codons. There is precedent for use of each of these strategies by other members of the order Mononegavirales. However, the concurrent use by BDV of such a diversity of strategies for the regulation of its gene expression is unique among known negative sense ssRNA viruses. In addition, as with viruses belonging to the family Orthomyxoviridae, BDV uses RNA splicing to generate some of its mRNAs. This represents another unique feature in the order Mononegavirales. BDV has one single surface glycoprotein gene (G) which is responsible for viral attachment and fusion upon endocytosis and endosome acidification. This pH-dependent fusogenic activity of G requires its post-translational cleavage by the cellular protease furin. Thus, BDV G expression and function appear to be a unique feature in negative sense ssRNA viruses, representing a combination of the strategies adopted by rhabdoviruses and paramyxoviruses.
Derivation of name
Borna refers to the city of Borna in Saxony, Germany, where many horses died in 1885 during an epidemic of a neurological disease, designated as Borna disease (BD), caused by the infectious agent presently known as Borna disease virus (BDV).
Further reading
2002. In: Carbone, K.M., Borna Disease Virus: Its Role in Neurobehavioral Disease. ASM Press, Washington DC.
Ludwig, H., Bode, L., Gosztonyi, G., 1988. Borna disease: a persistent virus infection of the central nervous system. Prog. Med. Virol. 35, 107–151.
Nowotny, N., Kolodziejek, J., Jehle, C.O., Suchy, A., Staeheli, P., Schwemmle, M., 2000. Isolation and characterization of a new subtype of Borna disease virus. J. Virol. 74, 5655–5668.
Perez, M., Sanchez, A., Cubitt, B., Rosario, D., de la Torre, J.C., 2003. A reverse genetics system for Borna disease virus. J. Gen. Virol. 84, 3099–3104.
Rudolph, M.G., Kraus, I., Dickmanns, A., Eickmann, M., Garten, W., Ficner, R., 2003. Crystal structure of the borna disease virus nucleoprotein. Structure. 11, 1219–1226.
Schneemann, A., Schneider, P.A., Lamb, R.A., Lipkin, W.I., 1995. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology. 210, 1–8.
Staeheli, P., Sauder, C., Hausmann, J., Ehrensperger, F., Schwemmle, M., 2000. Epidemiology of Borna disease virus. J. Gen. Virol. 81, 2123–2135.
Contributed by
In reproducing this chapter in edited form from the Eighth ICTV Report, the editors gratefully acknowledge the contribution of the authors, Schwemmle, M., Carbone, K.M., Tomonago, K., Nowotny, N. and Garten, W.
Figures
Figure 1 Negative contrast electron micrograph of a particle of Borna disease virus. The bar represents 100 nm.
(Courtesy of Dr M. Eickmann.)
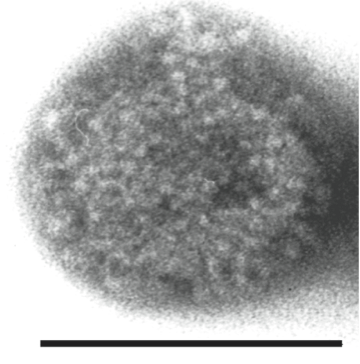
Figure 2 Genomic organization and transcriptional map of an isolate of Borna disease virus. ORFs are represented by boxes at the top. The location of transcription initiation and transcription termination sites are indicated by S and E, respectively. Positions of introns: I (nt 19322025), II (nt 24103703), and III (nt 24104559) are indicated. For details see text.