Family: Yadokariviridae
Yukiyo Sato, Subha Das, Leonardo Velasco, Massimo Turina, Hideki Osaki, Ioly Kotta-Loizou, Robert H. A. Coutts, Hideki Kondo, Sead Sabanadzovic and Nobuhiro Suzuki
The citation for this ICTV Report chapter is the summary published as Sato et al., (2023):
ICTV Virus Taxonomy Profile: Yadokariviridae 2023, Journal of General Virology 104:001826
Corresponding author: Nobuhiro Suzuki ([email protected])
Edited by: Sead Sabanadzovic and Stuart Siddell
Posted: December 2022
Summary
Yadokariviridae is a family of capsidless non-segmented positive-sense (+) RNA viruses in the order Yadokarivirales (Table 1.Yadokariviridae). The family includes the genera Alphayadokarivirus and Betayadokarivirus. Yadokarivirids lack their own capsid protein but hijack the capsids from phylogenetically distant double-stranded RNA (dsRNA) viruses on which yadokarivirids rely for their viability. Heterocapsids, which include the yadokarivirid RNA-directed RNA polymerase (RdRP) and replicative form dsRNA, are the likely replication site of yadokarivirids. Nevertheless, yadokarivirids show phylogenetic affinity to positive-sense (+) RNA viruses in the phylum Pisuviricota. Thus, yadokarivirids are evolutionary unique viruses that may have switched their replication site from host membranous structures to the heterocapsid, as with a dsRNA virus. While the known capsid donor dsRNA viruses span at least five diverse families, each yadokarivirid relies on a distinct species of dsRNA virus. Yadokarivirids play a complex role in the tripartite interaction with their capsid donor viruses and host fungi.
Table 1. Yadokariviridae. Characteristics of members of the family Yadokariviridae
|
Characteristic |
Description |
|
Example |
yado-kari virus 1 (LC006253), species Alphayadokarivirus ichibani genus Alphayadokarivirus |
|
Virion |
Capsidless per se. Trans-encapsidated into non-enveloped spherical virions, 33–50 nm in diameter, encoded by phylogenetically distant dsRNA viruses. |
|
Genome |
Non-segmented linear (+) RNA of 3.6–6.3 kb. |
|
Replication |
Assumed to replicate inside the heterocapsids encoded by a dsRNA virus. |
|
Translation |
From a genomic RNA serving as a polyprotein-encoding monocistronic or bicistronic mRNA with or without a poly(A) tail. |
|
Host range |
Fungi and possibly oomycetes. |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, order Yadokarivirales; The family includes two genera (Alphayadokarivirus and Betayadokarivirus) accommodating six and four species, respectively. |
Virion
Morphology
Yadokarivirids (viruses that are members of the family Yadokariviridae) lack their own capsid protein (CP). Instead, they are trans-encapsidated by specific dsRNA viruses (Table 2.Yadokariviridae, Figure 1.Yadokariviridae). This property of hetero-encapsidation gives rise to the name Yadokariviridae, derived from “yadokari”, which in Japanese means “room borrower”. The capsid donor (or “partner”) dsRNA viruses span at least five families in the order Ghabrivirales, a taxon that includes toti-like dsRNA viruses (Sato et al., 2022). The trans-encapsidated yadokarivirids are infectious in the presence of their partner dsRNA viruses (Zhang et al., 2016, Sato et al., 2022). The virions of trans-encapsidated yadokarivirids show non-enveloped spherical forms 33–50 nm in diameter, apparently identical to the virion size of their respective capsid donors.
Physicochemical and physical properties
The fractionation of mixed virus particles of yado-kari virus 1 (YkV1) and its partner an unclassified dsRNA virus (yado-nushi virus [YnV1]) by caesium chloride (CsCl) density gradient centrifugation reveals (Das et al., 2021) particle fractions containing YkV1 dsRNA and RdRP peaking at 1.395 g/cm3, while fractions containing both YkV1 and YnV1 dsRNAs peak at 1.408–1.496 cm3. It is concluded that YkV1 heterocapsids packaging YkV1 dsRNA and RdRP exist in infected mycelia. However, it remains unknown whether other types of particles, e.g., those encasing both YkV1 and YnV1 dsRNA, are produced, or how many dsRNA molecules are encased in one single particle.
Nucleic acid
The heterocapsids encase replicative form dsRNA of yadokarivirids. The ratio of encapsidated dsRNA of yadokarivirids to that of capsid donors in total virus particles varies depending on specific yadokarivid-donor virus combination. For example, the ratio of YkV1 dsRNA to its partner dsRNA is approximately one, whereas yado-kari virus 3 (YkV3) and its donor Rosellinia necatrix megabirnavirus 3 (family Megabirnaviridae) and for yado-kari virus 4a (YkV4a) and its donor the unclassified virus Rosellinia necatrix megatotivirus 1a, the ratios are approximately 0.1–0.2 and 0.6–0.7, respectively (Zhang et al., 2016, Sato et al., 2022).
Proteins
Virions encase yadokarivirid-encoded RdRP in addition to their dsRNA (Figure 1.Yadokariviridae) (Zhang et al., 2016, Jia et al., 2022). Reverse genetics analysis of YkV1 reveals that the RdRP of YkV1 is essential for its replication (Das et al., 2021). The RdRP of capsid donor viruses, expressed as the CP-RdRP fusion product or as a separate RdRP, may also be encased in the heterocapsids (Figure 1.Yadokariviridae).
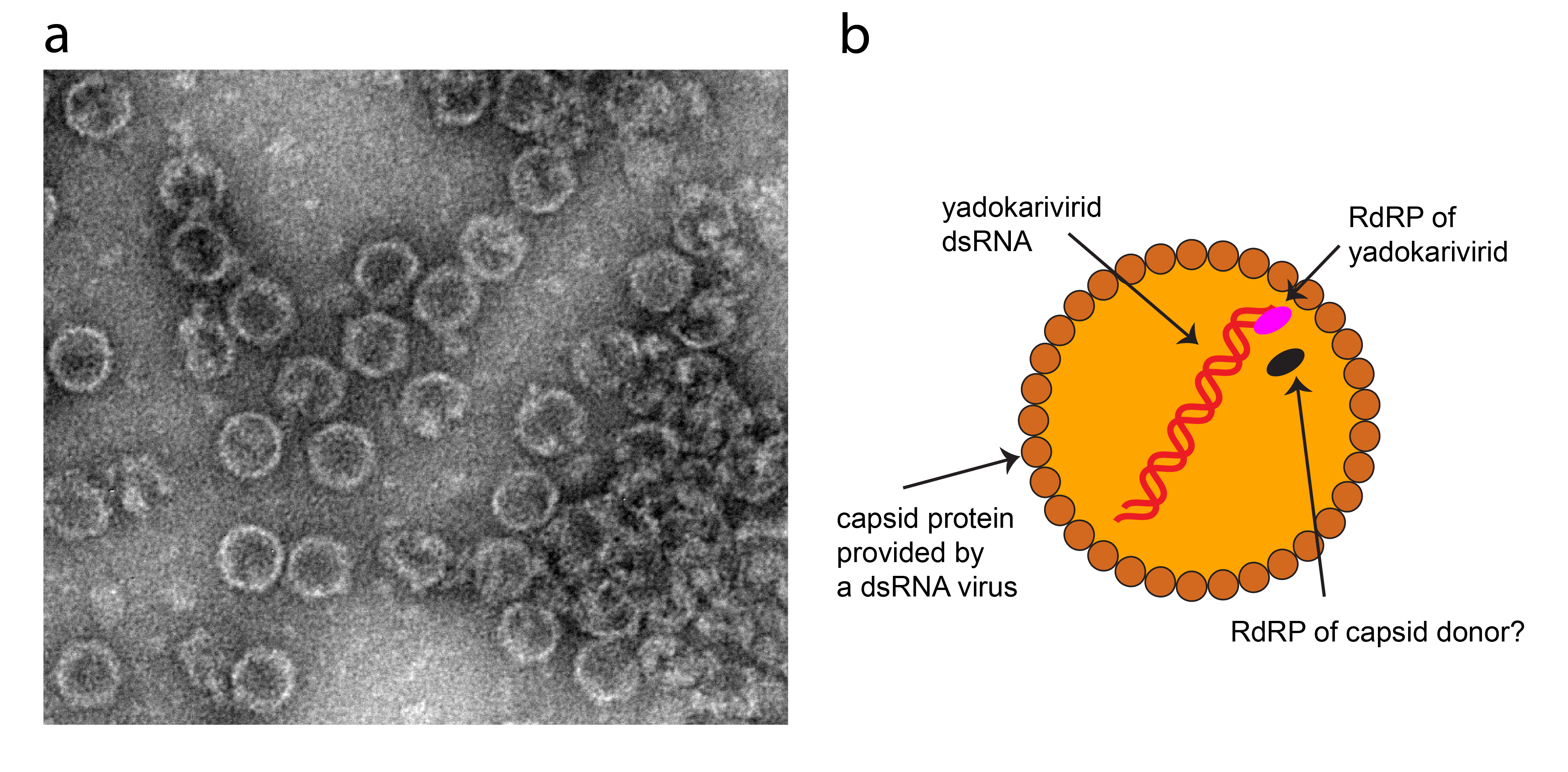 |
|
Figure 1. Yadokariviridae. (a) Transmission electron micrograph of negatively-stained virions of yado-kari virus 1 and its capsid donor the unclassified yado-nushi virus 1, prepared as previously described (Zhang et al., 2016). (b) Illustration of a hypothesized cross-section of the yadokarivirid virion. RdRP: RNA-directed RNA polymerase; CP: capsid protein. Whether the RdRP of a capsid donor is co-packaged along with yadokarivirid RNA and RdRP remains unknown. |
Table 2.Yadokariviridae. Some properties of yadokarivirids and their capsid donor viruses
|
Yadokarivirids |
Capsid donor dsRNA virus |
Reference |
|||||
|
Genus |
Full name (Abbreviation) |
Genome |
Full name (Abbreviation) |
Taxon |
CP mass |
Virion diameter (nm) |
|
|
Alphayadokarivirus |
yado-kari virus 1 (YkV1) |
6310 |
yado-nushi virus 1 (YnV1) |
Unclassified |
120 |
~40 |
|
|
Alphayadokarivirus |
Aspergillus foetidus slow virus 2 (AfSV2) |
3634 |
Aspergillus foetidus slow virus 1 (AfSV1) |
Family Totiviridae, |
78 |
33-37 |
|
|
Betayadokarivirus |
yado-kari virus 3 (YkV3) |
5681 |
Rosellinia necatrix megabirnavirus 3 (RnMBV3) |
Family Megabirnaviridae, |
131 |
~50 |
|
|
Betayadokarivirus |
yado-kari virus 4a (YkV4a) |
5343 |
Rosellinia necatrix megatotivirus 1 (RnMTV1) |
Unclassified |
130 |
~50 |
|
|
Betayadokarivirus |
yado-kari virus 4b (YkV4b) |
5349 |
Rosellinia necatrix megatotivirus 1a, 1b, & 1c (RnMTV1a, 1b, & 1c) |
Unclassified |
130 |
~50 |
|
|
Unclassified |
Sclerotinia sclerotiorum yadokarivirus 1 (SsYkV1) |
5256 |
Sclerotinia sclerotiorum botybirnavirus 3 (SsBV3) |
Genus Botybirnavirus |
~120 |
ND |
|
ND: not described.
Genome organization and replication
All known yadokarivirids have a non-segmented (+) RNA genome. Different yadokarivirids show similar yet distinct genome organization, coding strategy and terminal sequences (Figure 2.Yadokariviridae). Most have a monocistronic genome encoding a polyprotein containing a 2A-like self-cleaving peptide. The N- or C-terminal parts of the polyprotein, respectively, form the mature RdRP and a smaller protein. Both proteins are required for the replication of YkV1 (Zhang et al., 2016, Das et al., 2021). The two members of the species Betayadokarivirus yonbani have a putative bicistronic genome and do not encode a 2A-like motif. The mechanism of expression of their downstream open reading frames (ORFs) is unknown. The 5′-terminal end nucleotide of some yadokarivirid genomes shows heterogeneity (Arjona-Lopez et al., 2018). The genomes of some yadokarivirids are polyadenylated at the 3′-termini (Kozlakidis et al., 2013b, Arjona-Lopez et al., 2018, Velasco et al., 2019, Jia et al., 2022), while those of others are not (Nerva et al., 2016, Zhang et al., 2016).
Experiments using infectious cDNA clones of yadokarivirids, including YkV1, suggests that the replication competency of yadokarivirids depends on the presence of their respective partner dsRNA viruses (Zhang et al., 2016, Sato et al., 2022). Yadokarivirids have been hypothesized to replicate inside the hetero-capsids provided by a donor virus as if they were true dsRNA viruses, despite their evolutionary relatedness to (+) RNA viruses (Hisano et al., 2017, Das and Suzuki 2021).
Sclerotinia sclerotiorum yadokarivirus 1 (SsYkV1), an unclassified virus in this family, appears to have both a circular and a linear form of the genomic RNA, the circular form being resistant to linear RNA-specific ribonuclease R (Jia et al., 2022). The presence, role, replication mechanism, and cellular localization of this circular RNA for other yadokarivirids remain unknown.
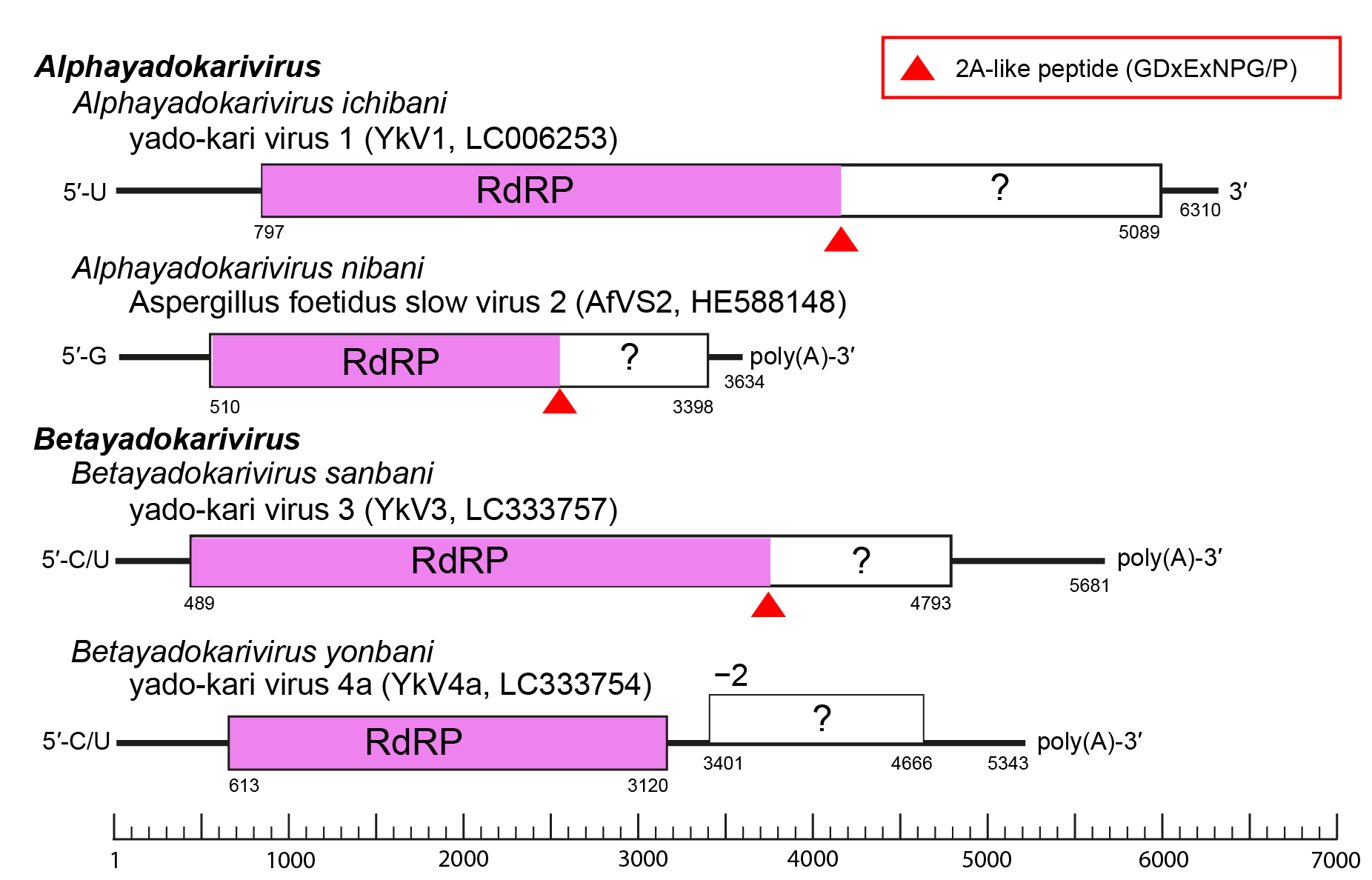 |
|
Figure 2. Yadokariviridae. Genome organization of two alphayadokariviruses and two betayadokariviruses. |
Biology
Yadokarivirids have been isolated from various ascomycetous fungi. In addition, an alphayadokarivirus (Plasmopara viticola lesion associated yadokari virus 1) has been reported from an environmental sample associated with a biotrophic unculturable oomycete, Plasmopara viticola (Chiapello et al., 2020). Yadokarivirids have been detected from hosts collected from several places in Asia (Japan and China) (Yaegashi et al., 2013, Osaki et al., 2016, Zhang et al., 2016, Jia et al., 2022), Europe (Spain and Italy) (Arjona-Lopez et al., 2018, Velasco et al., 2019, Chiapello et al., 2020) and the Middle East (Turkey) (Sahin et al., 2021). While the trans-encapsidation of yadokarivirids by capsids encoded by specific dsRNA viruses has been established for some viruses (Table 2.Yadokariviridae), all known yadokarivirids have been found to coinfect single host fungi together with diverse dsRNA viruses that could act as partners (Sato et al., 2022). Whether yadokarivirids other than those listed in Table 2.Yadokariviridae all rely on the co-infecting dsRNA viruses for viability, or in some cases can replicate independently, remains to be determined.
The partnership between yadokarivirids and their partner dsRNA viruses appears to be very specific. Partner swapping experiments with YkV1 (species Alphayadokarivirus ichibani), YkV3 (species Betayadokarivirus sanbani), YkV4a, and YkV4b (both members of the species Betayadokarivirus yobani) show that each yadokarivirid partners only with a specific dsRNA virus (Table 2.Yadokariviridae) (Sato et al., 2022). The partner pairs can be swapped only between different strains within a single species, but not between members of different species. It will be a future challenge to investigate the determinant(s) of partnership specificity. Although each yadokarivirid has species-level partner specificity, yadokarivirids as a whole can partner with diverse dsRNA viruses belonging to two families; one genus unassigned to a family and two additional unclassified groups in the order Ghabrivirales (Table 2.Yadokariviridae). Interestingly, the partnership between yadokarivirids and donor dsRNA viruses is incongruent with their phylogenetic relationships. For example, the betayadokarivirus YkV3 partners with the megabirnavirus RnMBV3, a virus belonging to the proposed suborder Alphatotivirinae, while another betayadokarivirus, YkV4, partners with RnMTV1, an unclassified member of another proposed suborder (Betatotivirinae), this proposed suborder also accommodating the unclassified donor virus YnV1, the partner of the alphayadokarivirus, YkV1 (Sato et al., 2022).
Yadokarivirids make complex symbiotic (mutualistic, commensal or harmful) relationships with the partner dsRNA viruses and host fungi (Sato et al., 2022). The partner dsRNA viruses and the host confer beneficial effects on yadokarivirids because they provide capsids or cellular environments for their replication, respectively. In turn, YkV1, for example, enhances the replication of its partner virus, while their coinfection induces a growth defect in the host fungus Rosellinia necatrix (Yaegashi et al., 2013, Zhang et al., 2016). Therefore, YkV1 seems to be beneficial to the partner virus but harmful to the host fungus. In contrast, the viruses YkV4a and YkV4b decrease the replication of their partner virus (Rosellinia necatrix megatotivirus 1a, RnMTV1a), which rescues the host R. necatrix from the growth inhibition caused by RnMTV1a (Sato et al., 2022). That is, YkV4a and YkV4b seem to be harmful to the partner virus but mutualistic to the host fungus. YkV3 shows no obvious effects on either its partner virus or the host R. necatrix (Sato et al., 2022), and so appears to be commensal to them both. Note that these effects are observed under experimental conditions on a synthetic growth medium. Since R. necatrix is a plant pathogen, it is of great interest to investigate the relationships yadokarivirids have with plants in this multi-layered interaction system. Other biological studies are limited to SsYkV1 which has no apparent impact on hypovirulence of the host phytopathogenic fungus Sclerotinia sclerotiorum induced by other co-infecting viruses (Osaki et al., 2016, Jia et al., 2022).
Antigenicity
Antibodies against the RdRP of the alphayadokarivirus YkV1 and the CP of its capsid donor (YnV1) are available (Zhang et al., 2016).
Derivation of names
Yadokariviridae: derived from the Japanese word “yadokari” (やどかり, 宿借) meaning “room borrower” or “hermit crab”.
Alpha and Beta prefixes are derived from the first and second letters of the Greek alphabet.
For both genera, species epithets are derived from the Japanese ordinal numbers (ichiban, niban, sanban, …) with a Latinized suffix (-i).
Genus demarcation criteria
· The phylogenetic position based on the amino acid sequence of RdRP
· The similarity of nucleotide or amino acid sequences compared with the existing members
Relationships within the family
Yadokarivirids are phylogenetically divided into two clades based on the RdRP amino acid sequence (Figure 3.Yadokariviridae), corresponding to the two genera Alphayadokarivirus and Betayadokarivirus. The identity of the RdRP polyprotein between members of the distinct species is 27–65% within the genus Alphayadokarivirus and 34–68% within the genus Betayadokarivirus, but only19–25% between members of different genera (see the 2021 ICTV taxonomy proposal for the order Yadokarivirales https://ictv.global/files/proposals/approved?fid=4894). There are no genus-specific differences in genome organization (e.g. the monocistronic or bicistronic nature; the presence or absence of poly(A) tail) or other traits.
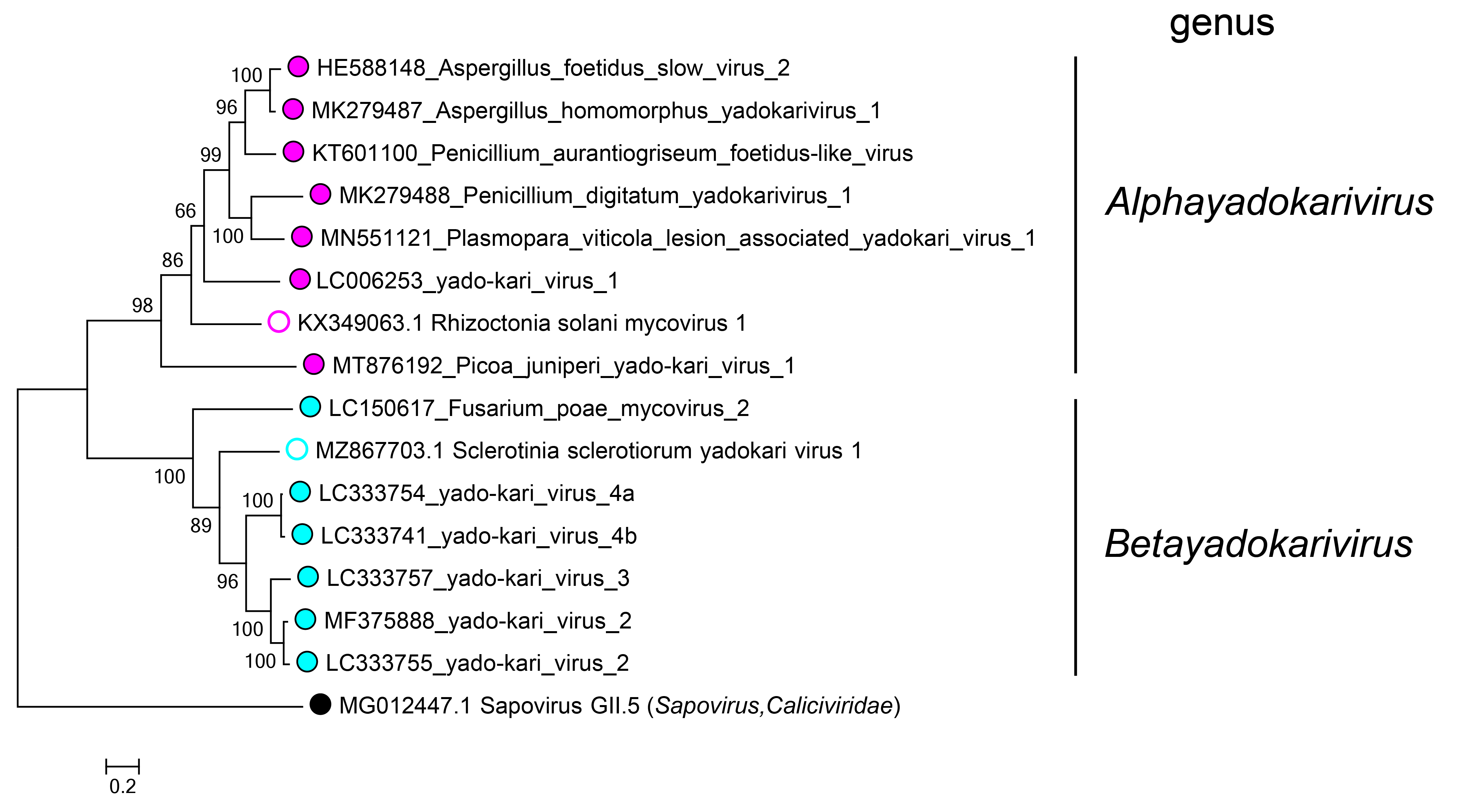 |
|
Figure 3.Yadokariviridae. Phylogenetic tree of members of the family Yadokariviridae. Alphayadokariviruses and betayadokariviruses are shown with solid magenta or cyan circles, respectively; unassigned viruses are labelled with open circles. A sapovirus in the family Caliciviridae was included as an outgroup. The phylogeny has previously been inferred by a maximum likelihood method based on multiple alignments of RdRP amino acid sequences (Sato et al., 2022). The values next to the branches indicate bootstrap probabilities in 1000 iterations. The phylogenetic tree was visualized in MEGA X (Kumar et al., 2018). |
Relationships with other taxa
Yadokariviridae is currently the only family in the order Yadokarivirales, both established in 2021. The RdRP of yadokarivirids shows homology to several groups of (+) RNA viruses in the phylum Pisuviricota (extended picornavirus supergroup). For instance, apart from other yadokarivirids, the RdRP of YkV1 shows the highest sequence identity by BLAST search to that of sapoviruses in the family Caliciviridae (Zhang et al., 2016). Based on their phylogenetic placement, yadokarivirids are regarded as (+) RNA viruses (Hisano et al., 2017). Their phylogenetic relation is distant from any specific order or class in the phylum Pisuviricota. These phylogenetic affinities and yadokarivirid nature led to the creation of the unique order Yadokarivirales in this phylum without assignment to a class (see the ICTV taxonomy proposal for the order Yadokarivirales, 2021, https://ictv.global/files/proposals/approved?fid=4894).

