Family: Hadakaviridae
Yukiyo Sato, Massimo Turina, Sotaro Chiba, Ryo Okada, Muhammad F. Bhatti, Ioly Kotta-Loizou, Robert H. A. Coutts, Hideki Kondo, Sead Sabanadzovic and Nobuhiro Suzuki
The citation for this ICTV Report chapter is the summary published as Sato et al., (2023):
ICTV Virus Taxonomy Profile: Hadakaviridae 2023, Journal of General Virology (2023) 104:001820
Corresponding author: Nobuhiro Suzuki ([email protected])
Edited by: Sead Sabanadzovic and Peter Simmonds
Posted: November 2022
Summary
Hadakaviridae is a family of capsidless positive-sense (+) single-stranded RNA viruses with a genome composed of ten or eleven segments (Table 1.Hadakaviridae). “Hadaka”, meaning “naked” in Japanese and used to name the virus family, describes their unusual capsidless natures. The lack of a capsid means that these viruses cannot be pelleted by conventional ultracentrifugation and their genome and replicative form double-stranded RNAs are susceptible to exogenously added ribonuclease in host tissue homogenates. Currently known hosts are ascomycetous filamentous fungi. Hadakavirids are most closely related to members of the family Polymycoviridae and show phylogenetic affinity to (+) RNA viruses in the phylum Pisuviricota (extended “picornavirus supergroup”).
Table 1. Hadakaviridae. Characteristics of members of the family Hadakaviridae
|
Characteristic |
Description |
|
Example |
hadaka virus 1 (LC519840–LC519850), species Hadakavirus nanga genus Hadakavirus |
|
Virion |
Capsidless (no known virions) |
|
Genome |
Multi-segmented genome of ten or eleven linear, positive-sense (+) RNA segments of 0.9–2.5 kb, comprising 14–15 kb in total |
|
Replication |
Double-stranded replicative forms accumulate abundantly in infected fungal hosts. The cellular replication site remains unknown. |
|
Translation |
From monocistronic non-polyadenylated genomic RNAs |
|
Host range |
Fungi |
|
Taxonomy |
Realm Riboviria; kingdom Orthornavirae, phylum Pisuviricota; The family includes one genus Hadakavirus accommodating one species |
Virion
Morphology
Hadakavirids (members of the family Hadakariviridae) have no true virion, and they cannot be purified by conventional virus purification methods (Sato et al., 2020b, Khan et al., 2021). Their infectious entity remains unknown.
Physicochemical and physical properties
Hadakavirids cannot be pelleted by ultracentrifugation at 100,000 × g for 1.5 h at 4°C, conditions that can normally be used to purify encapsidated viruses independently of their virion size or morphology (Sato et al., 2020b).
Nucleic acid
The genome of hadaka virus 1 (HadV1) consists of ten or eleven positive-sense (+) RNA segments (Sato et al., 2020b, Khan et al., 2021). Colletotrichum fructicola RNA virus 1 (CfRV1), an unassigned member of the family Hadakaviridae, has only seven genomic segments (Fu et al., 2022). Double-stranded (ds) RNA replicative forms accumulate abundantly in host fungal mycelia. Both, the hadakavirid genomic (+) RNA and dsRNAs, are susceptible to ribonuclease A (exogenously added) in host tissue homogenates buffered with a neutral phosphate (Sato et al., 2020b, Khan et al., 2021). How these RNAs are protected in vivo from endogenous host nucleases remains to be determined.
Proteins
No structural proteins have been detected experimentally and/or bioinformatically (Sato et al., 2020b, Khan et al., 2021). The hypothetical proteins encoded by the hadakavirid genome show no homology to known viral structural proteins.
Lipids
CfRV1 appears to be associated with the formation of giant vesicles (196–559 nm) containing smaller vesicles (25–40 nm) in host fungal cells (Fu et al., 2022). These vesicles appear to be lipid -layered. However, it remains to be determined whether these vesicles contain viral components.
Genome organization and replication
The exemplar virus isolate (HadV1-7n) has an eleven-segmented (+) RNA genome, each containing a single open reading frame (ORF) (Figure 1.Hadakaviridae), while hadaka virus 1-1NL (HadV1-1NL) has ten monocistronic segments (Sato et al., 2020b, Khan et al., 2021). All genome segments start with the conserved three nucleotides 5′-CGUand end with CCA-3′ or CCG-3′. Exceptions to this rule are RNA7 of HadV1-7n and RNA6 of HadV1-1NL, which have the termini 5′-CGU and GGG-3′. Genomic segments are vertically transmitted through asexual spores in an all-or-none fashion. The largest three genomic segments, RNAs1–3, encode proteins homologous to those encoded by dsRNAs1–3 of members of the family Polymycoviridae, namely an RNA-dependent RNA polymerase (RdRP), a hypothetical protein with unknown functions, and a putative methyltransferase (MTR). A unique genomic segment, RNA8, of HadV1-7n encodes a short C2H2-type zinc finger protein. This genomic segment is missing for HadV1-1NL (Khan et al., 2021). Hypothetical proteins encoded by the remaining genomic segments have no significant homology to other known proteins. The other potential member of the Hadakaviridae, CfRV1, has fewer (seven) genomic segments (Fu et al., 2022) with the three largest also encoding proteins homologous to those of polymycovirids. Most CfRV1 genomic segments possess the conserved terminal sequences 5′-CGU and CCN-3′.
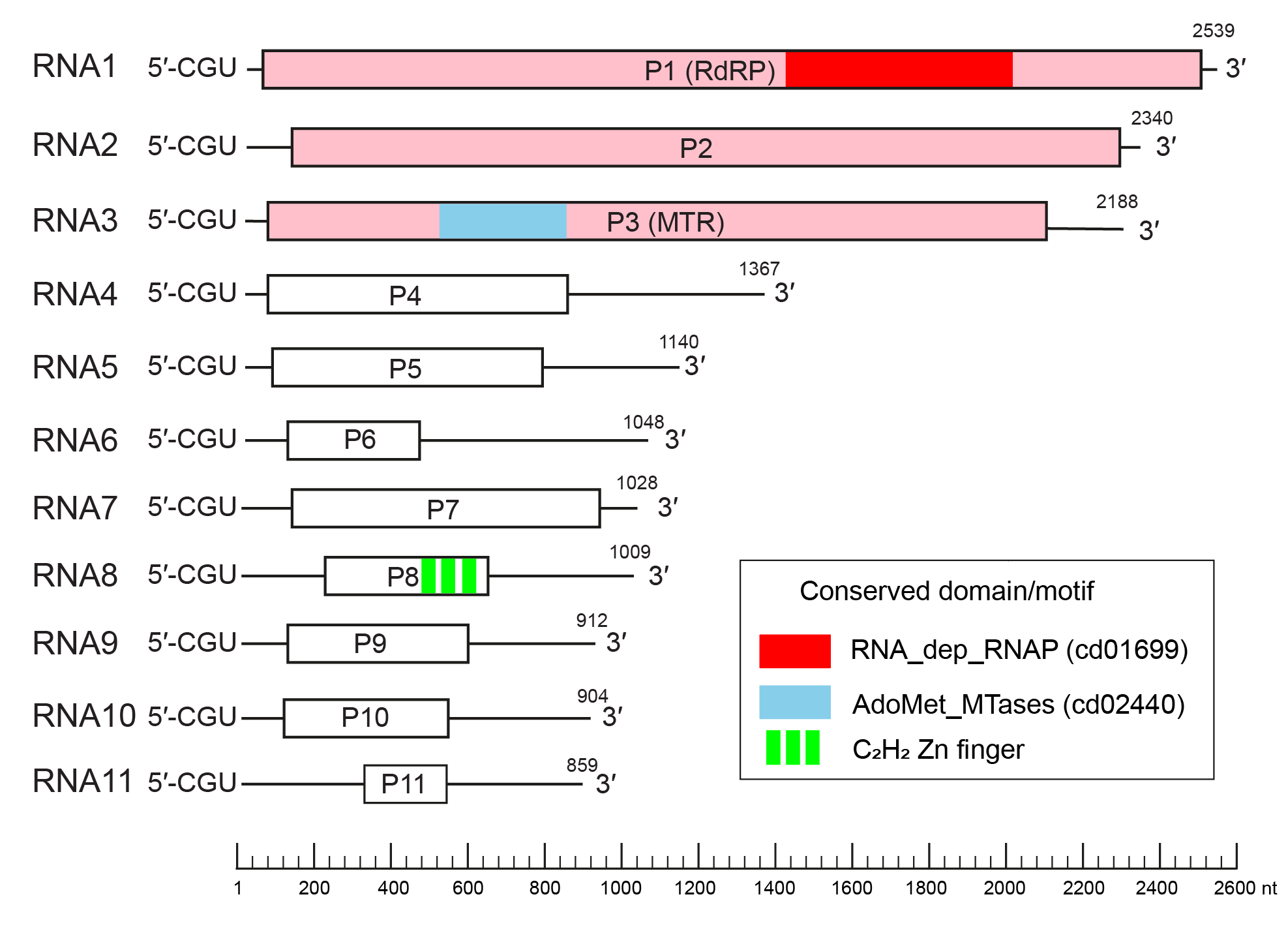 |
|
Figure 1. Hadakaviridae. Genome organization of hadaka virus 1 isolate 7n (LC519840–50). Open reading frames are indicated by boxes. The three genes homologous between hadakavirids and polymycovirids are coloured pink: P1 (RdRP - RNA-dependent RNA polymerase), P2 (unknown function) and P3 (MTR- methyltransferase). Other colored regions indicate conserved domains or motifs identified previously (Sato et al., 2020b). The conserved protein domains and motifs were detected from the Conserved Domain Database (Lu et al., 2020). |
Biology
Known hosts for the hadakavirids are ascomycetous filamentous fungi. HadV1-7n and -1NL were isolated from Fusarium oxysporum and F. nygamai, respectively, in Pakistan (Sato et al., 2020b, Khan et al., 2021). Neither of these HadV1 isolates have any apparent effect on the host on potato dextrose agar (PDA) media. CfRV1 was discovered from Colletotrichum fructicola in China (Fu et al., 2022). CfRV1 causes a mild growth reduction of the host on PDA medium and an infection delay on harvested pear fruits. No experimental method for infecting the host with extracellular hadakavirids has been developed.
Antigenicity
No antibody against any proteins of hadakavirids has been reported.
Derivation of names
Hadakaviridae: derived from Hadaka (裸、はだか), a Japanese word meaning “naked”.
Hadakavirus nanga: derived from nanga (ننگا), an Urdu word meaning “naked”, Urdu being the official language of Pakistan where HadV1-7n and HadV1-1NL were isolated.
Genus demarcation criteria
No genus demarcation criteria have been defined since the family Hadakaviridae currently consists of one single genus.
Relationships within the family
The family includes a single genus Hadakavirus which accommodates one species, Hadakavirus nanga, with two well-characterized virus isolates HadV1-7n and -1NL (Sato et al., 2020b, Khan et al., 2021). CfRV1 is phylogenetically closer to HadV1-7n and HadV1-1NL than to polymycovirids (Figure 2.Hadakaviridae) (Fu et al., 2022).
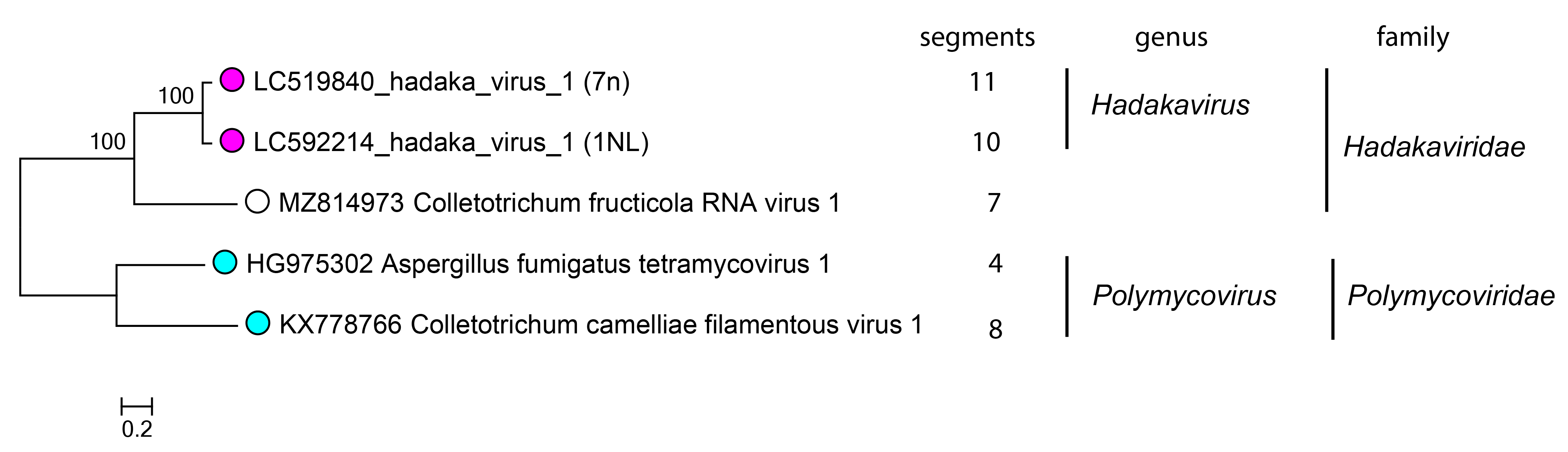 |
|
Figure 2.Hadakaviridae. Phylogenetic tree of hadakavirids and related viruses. Members of the same genus have tips labelled with the magenta (Hadakvirus) or cyan (Polymycovirus – used as an outgroup); an unassigned virus as an open circle. The maximum likelihood tree was generated based on the amino acid sequence alignment of RdRP. Multiple sequence alignment was performed by the L-INS-i method in MAFFT v7.490 (Katoh and Standley 2013) and trimmed by trimAl v1.4.rev15 with the default parameter settings (Capella-Gutiérrez et al., 2009). The phylogeny was inferred by IQ-TREE v.2.0.3 with the best fit model LG+G4 (Minh et al., 2020). The confidence was analyzed by the ultrafast bootstrap method in 1000 replicates. The tree was constructed using MEGA X (Kumar et al., 2018). |
Relationships with other taxa
Hadakavirids are most closely related to members of the family Polymycoviridae (Kotta-Loizou et al., 2022), based on phylogenetic analyses of the three conserved proteins (Hadaka/Polymyco-P1, P2 and P3 in Figure 2.Hadakaviridae) (Sato et al., 2020b, Khan et al., 2021, Fu et al., 2022). The largest difference between hadakavirids and polymycovirids is the absence or presence of the genome segment encoding a proline-alanine-serine-rich protein (PASrp). PASrp is known to physically associate with the dsRNA genome of polymycovirids (Kanhayuwa et al., 2015, Sato et al., 2020a) and so gives polymycovirids an intermediate position between encapsidated and capsidless RNA viruses. Unlike polymycovirids and conventionally encapsidated viruses, HadV1 cannot be pelleted by ultracentrifugation and is susceptible to exogenously added ribonuclease A in host tissue homogenates (Sato et al., 2020a, Sato et al., 2020b).
Based on the conserved RdRP motifs, hadakavirids show phylogenetic affinity to (+) RNA viruses, such as the members of the family Astroviridae and Caliciviridae, and is therefore classified in the phylum Pisuviricota (extended “picornavirus supergroup”) (Sato et al., 2020b, Fu et al., 2022) along with other capsidless (+) RNA virus taxa such as the family Hypoviridae (Suzuki et al., 2018). The catalytic core amino acid residues of the RdRPs of hadakavirids and polymycovirids are “GDNQ”, which is a hallmark of some negative-sense RNA viruses in the order Mononegavirales, whereas the enzymes encoded by most members of the phylum Pisuviricota are characterized by a “GDD” triplet.

