Family: Tosoviridae
Yuri I. Wolf, Mart Krupovic, Jens H. Kuhn and Eugene V. Koonin
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Eugene V. Koonin ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: October 2023
Summary
Tosoviridae is a family of negative-sense RNA viruses with genomes of about 12.3 kb (Table 1.Tosoviridae). These viruses have been found in trionychid and emydid testudines (softshell turtles and cooters) in Florida, USA. The family includes a single genus with one species for one virus. The tosovirid genome consists of two segments, each with two open reading frames (ORFs) in ambisense orientation. The small (S) segment encodes a nucleoprotein (NP) and a glycoprotein precursor (GPC), and the large (L) segment encodes a large (L) protein with an RNA-directed RNA polymerase (RdRP) domain and a zinc-binding (Z) protein.
Table 1.Tosoviridae Characteristics of members of the family Tosoviridae
| Characteristic | Description | |
| Example | turtle fraservirus 1 (S segment: MZ458542; L segment: MZ458543), species Fraservirus testudinis, genus Fraservirus | |
| Virion | Enveloped round particles of 110–125 nm | |
| Genome | About 12.3 kb of bi-segmented negative-sense RNA | |
| Replication | Unknown | |
| Translation | Unknown | |
| Host range | Emydid and trionychid turtles | |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Hareavirales; the family includes one genus and one species. | |
Virion
Morphology
Tosovirids produce enveloped round to pleomorphic particles that are 110–125 nm in diameter with prominent surface peplomers (Waltzek et al., 2022) (Figure 1.Tosoviridae).
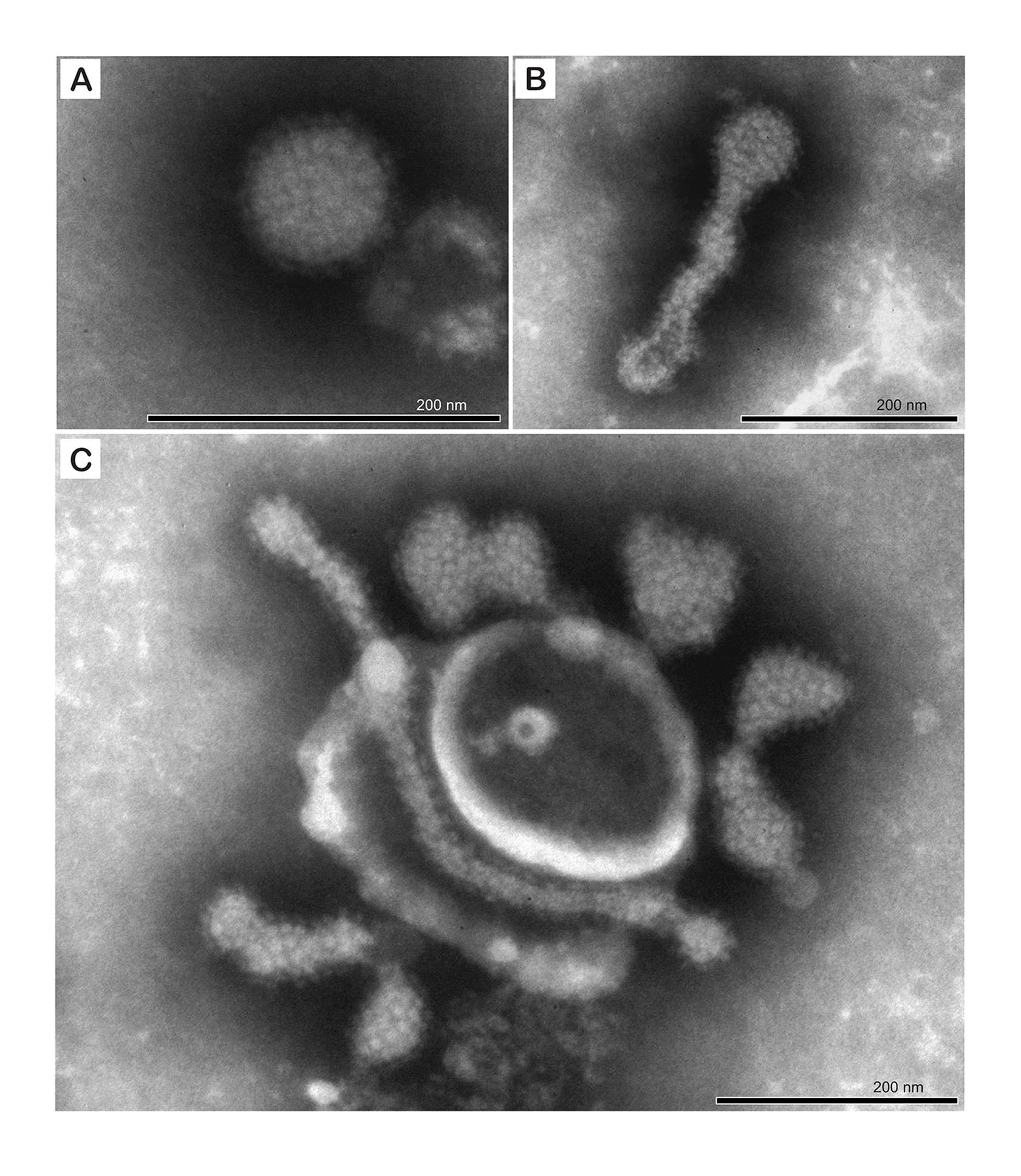 |
| Figure 1.Tosoviridae. Negative-stain electron photomicrograph illustrating the ultrastructural features of the Turtle fraservirus 1 (TFV1) virion. A) spherical particle with prominent glycoprotein spikes. B and C) pleomorphic particles including elongated forms. Scale bars = 200 nm (reproduced from Waltzek et al., 2022). |
Nucleic acid
Tosovirids have two segments (S and L) of linear negative-sense RNA with a total length of about 12.3 kb (S segment: about 5.7 kb; L segment: 6.6 kb) (Waltzek et al., 2022).
Genome organization and replication
Viruses of the family Tosoviridae have bi-segmented genomes, with each segment possessing two ORFs in ambisense orientation that are separated by non-coding intergenic regions (IGRs) (Figure 2.Tosoviridae). The IGRs form one or more energetically stable stem–loop (hairpin) structures that function in structure-dependent transcription termination and in virion assembly and budding. The S segment encodes an NP and a GPC; the L segment encodes an L protein containing an RdRP domain, and a Z protein (Waltzek et al., 2022). The RdRP is only distantly related to the RdRPs of all other currently classified negarnaviricots, whereas the GPC is homologous to that encoded by hantavirid actinoviruses (Bunyavirales) (Waltzek et al., 2022). The replication cycle of tosovirids remains to be elucidated.
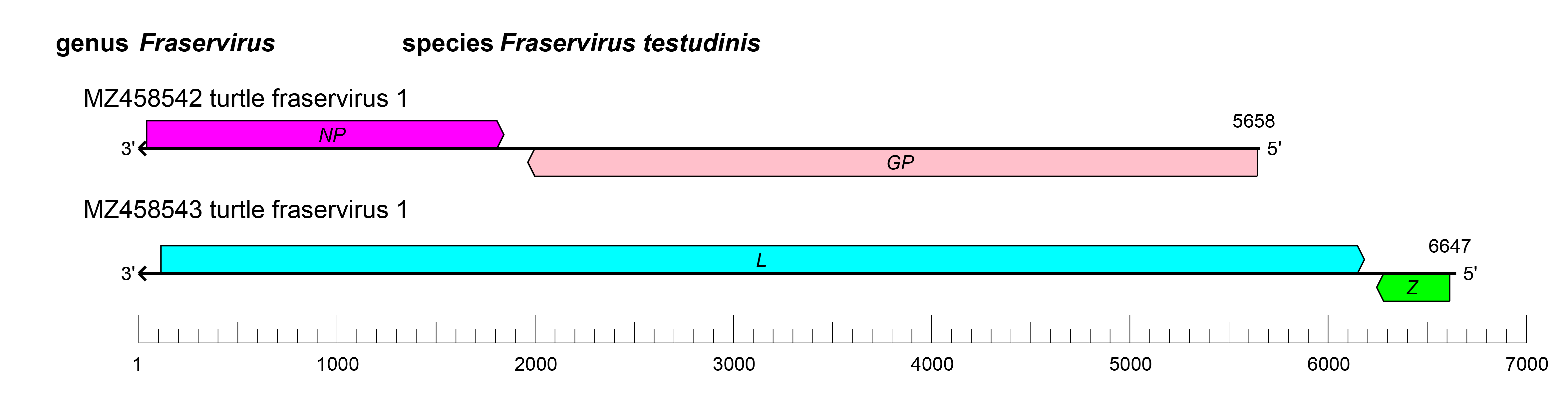 |
| Figure 2.Tosoviridae. Genome length, organization, and position of open reading frames (ORFs) of turtle fraservirus 1. ORFs are indicated as boxes, colored according to the predicted protein function (GPC, glycoprotein precursor gene; NP, nucleoprotein gene; L, large protein gene; Z, zinc-binding protein gene). |
Biology
The only classified tosovirid, turtle fraservirus 1 (TFV1), was first isolated from diseased Florida softshell turtles (trionychid Apalone ferox (Schneider, 1783)) and later detected in diseased Florida red-bellied cooters (emydid Pseudemys nelson Carr, 1938) and peninsula cooters (emydid Pseudemys peninsularis Carr, 1938) in Florida, USA (Waltzek et al., 2022).
In the turtles in which it was detected, TFV1 caused neurological signs and genital and oral ulcerations. Pathologic findings included heterophilic/histiocytic meningoencephalitis and multi-organ vasculitis (Waltzek et al., 2022).
An unclassified tosovirid has been found in New Zealand longfin eels (anguillid Anguilla dieffenbachii J. E. Gray, 1842) and shortfin eels (Anguilla australis J. Richardson, 1841) in Lake Te Wapu, New Zealand (Grimwood et al., 2023).
Derivation of names
Fraservirus: from TVF1 co-describer William A. Fraser
testudinis: from host order Testudines
Tosoviridae: from Tosohatchee Wildlife Management Area, Florida, USA
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Not applicable (the only genus includes only a single species).
Relationships within the family
One other L segment sequence is available (OM471797, turtle fraservirus 1 strain TFV1/2007), this being 99.26% identical to that of the exemplar isolate (MZ458543).
Relationships with other taxa
Viruses in the family Tosoviridae have a similar genome organization to viruses assigned to the very distantly related bunyaviral Arenaviridae (Waltzek et al., 2022, Radoshitzky et al., 2023). The phylogenetic relationship of TVF1 to other viruses in the phylum Negarnaviricota is shown in Figure 3.Tosoviridae.
 |
| Figure 3.Tosoviridae. Phylogenetic relationships of turtle fraservirus 1 with viruses in the phylum Negarnaviricota. The list of complete exemplar genomes of Negarnaviricota was obtained from the ICTV Virus Metadata Resource (VMR) datasheet v.38.1 (https://ictv.global/vmr). RdRP core domains were identified by running a PSI-BLAST search against either the protein coding gene complement or six-frame translation of the virus RNA sequences using the 143 negarnaviricot profiles (Neri et al., 2022) and a tosovirid profile (Waltzek et al., 2022). RdRP core fragments were clustered using MMSEQS2 with the sequence similarity threshold of 0.3; sequences in each cluster were aligned using MUSCLE5. A consensus sequence was derived from each alignment; consensus sequences were aligned using MUSCLE5, then each consensus amino acid was expanded into an alignment column (Neri et al., 2022). An approximate ML phylogenetic tree was reconstructed from this alignment of 1,122 negarnaviricot RdRP core sequences using FastTree (WAG evolutionary model, gamma-distributed site rates). A representative sequence (one, closest to the family root) was selected for each of the 36 negarnaviricot families and the full tree was trimmed to these 36 representative leaves. |
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Chatham eel tosovirus | L: OR270069; S: OR270088*; S: OR270089* | (Grimwood et al., 2023) |
Virus names and virus abbreviations are not official ICTV designations.
* Incomplete genome

