Family: Sphaerolipoviridae
Tatiana A. Demina, Mike Dyall-Smith, Matti Jalasvuori, Shishen Du, Hanna M. Oksanen
The citation for this ICTV Report chapter is the summary that has been published as Demina et al., (2023):
ICTV Virus Taxonomy Profile: Sphaerolipoviridae 2023, Journal of General Virology 2023;104:001830
Corresponding author: Hanna M. Oksanen (hanna.oksanen@helsinki.fi)
Edited by: Mart Krupovic and Stuart Siddell
Posted: February 2023
Summary
Members of the family Sphaerolipoviridae have non-enveloped tailless icosahedral virions with an internal membrane (Table 1.Sphaerolipoviridae). The genome is a linear double-stranded DNA of about 30 kbp with inverted terminal repeats and covalently bound terminal proteins. The capsid has a pseudo triangulation T = 28 dextro symmetry and is built of two major capsid protein types. The five-fold vertices have penton protein and spike complexes. Several membrane protein types are embedded in the internal membrane. All viruses infect halophilic archaea in the class Halobacteria (phylum Euryarchaeota). Sphaerolipoviruses have a narrow host range and a lytic life cycle.
Table 1.Sphaerolipoviridae. Characteristics of members of the family Sphaerolipoviridae.
| Characteristic | Description |
| Example | Haloarcula californiae icosahedral virus 1 (HCIV-1; KT809302), species Alphasphaerolipovirus viikkii |
| Virion | Non-enveloped, tailless icosahedral virion with an internal lipid membrane, diameter 80 nm, capsid is pseudo T = 28 dextro, two types of major capsid protein, horn-shaped or propeller-shaped five-fold vertex spike complexes, membrane associated proteins |
| Genome | Linear dsDNA, 28–31 kbp, with inverted terminal repeats and terminal proteins attached |
| Replication | Possibly protein-primed |
| Translation | Prokaryotic translation using viral mRNA and host ribosomes |
| Host range | Archaea, euryarchaeal Haloarcula and Halorubrum strains |
| Taxonomy | Realm Singelaviria, kingdom Helvetiavirae, phylum Dividoviricota, class Laserviricetes, order Halopanivirales; one genus (Alphasphaerolipovirus) with four species |
Virion
Morphology
Sphaerolipoviruses have tailless icosahedral virions with an internal protein-rich membrane vesicle (Figure 1.Sphaerolipoviridae) (Bamford et al., 2005, Porter et al., 2005, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016). The virion is typically about 80 nm in diameter. The capsid has a pseudo T = 28 dextro triangulation number (Jäälinoja et al., 2008, Gil-Carton et al., 2015, Santos-Pérez et al., 2019). There are two types of major capsid protein (MCP) with a vertical single jelly-roll fold: protein VP7 (Haloarcula californiae icosahedral virus 1 [HCIV-1] small MCP, a single jelly-roll protein) and protein VP4 (HCIV-1 large MCP, two single jelly-roll domains on top of each other). The two MCPs form pseudo-hexameric capsomers with either two or three towers (Figure 1.Sphaerolipoviridae). The capsid lattice is built of VP4-VP4 homodimers and VP4-VP7 heterodimers (Gil-Carton et al., 2015, Santos-Pérez et al., 2019). There are two different scaffolding protein complexes underneath the capsomers guiding the position of the capsomers on the membrane vesicle (Gil-Carton et al., 2015). The five-fold vertices are occupied by a penton protein with a single jelly-roll fold (HCIV-1 VP9 protein), forming the binding position for the spike complex, which serves in host recognition. Vertex complexes are either horn-shaped or propeller-shaped (Jäälinoja et al., 2008, Gil-Carton et al., 2015, Santos-Pérez et al., 2019). Spike complexes are multi-protein complexes e.g. proteins VP3, VP6, VP2 in the horn-shaped spike complex of HCIV-1 and SH1 virus (SH1) (Figure 1.Sphaerolipoviridae panels a and c), and VP17 and VP2 in the propeller-shaped spike complex of Haloarcula hispanica icosahedral virus 2 (HHIV-2) (Kivelä et al., 2006, Jaakkola et al., 2012, Santos-Pérez et al., 2019).
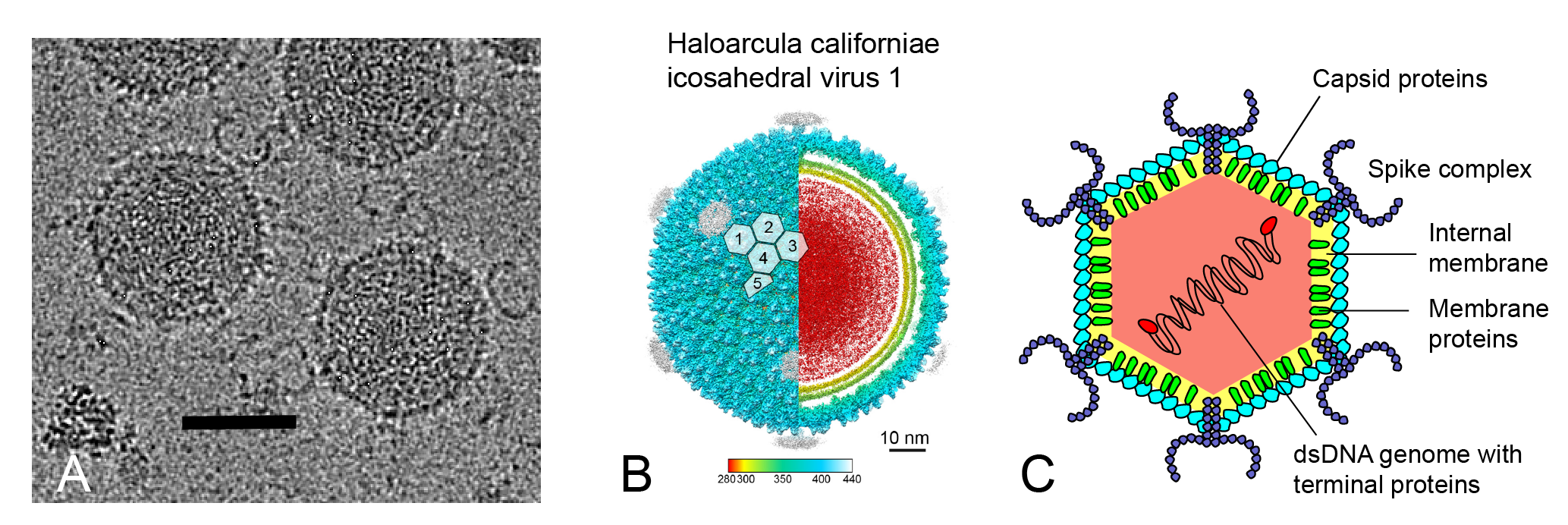 |
| Figure 1.Sphaerolipoviridae. Morphology of sphaerolipovirus virions. (a) Cryo-electron micrograph of HCIV-1 virions, scale bar: 50 nm, credit: Nicola Abrescia lab, CICbioGUNE, Spain. (b) Overall view of the cryo-electron microscopy density map of HCIV-1, color-coded by distance from the center. HCIV-1 is rendered to display the capsid shell (left-half) and the particle interior (right-half), genome in red and inner and outer membrane leaflets in yellow and yellow-lime (vertex complexes have been omitted). White-transparent hexagons on top of the capsid densities mark the capsomers (numbered 1–5) forming the icosahedral asymmetric unit. Reprinted under Creative Commons 4.0 from (Santos-Pérez et al., 2019). (c) Schematic model of the sphaerolipovirus HCIV-1 virion, modified from (Demina et al., 2017). |
Physicochemical and physical properties
Sphaerolipoviruses are typically stable at high ionic strength for several weeks at 5 °C. HCIV-1 and HHIV-2 can tolerate low ionic strength conditions (Jaakkola et al., 2012, Demina et al., 2016), while SH1 infectivity is dependent on Mg2+ and Ca2+ ions (Porter et al., 2005, Jäälinoja et al., 2008). Typically, sphaerolipoviruses are stable at temperatures below 50–60 °C and are more stable at neutral or slightly alkaline pH (pH 7–9) than in acidic conditions (pH ≤6) (Porter et al., 2005, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016). The virion buoyant density in CsCl is 1.28–1.33 g/ml (Porter et al., 2005, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016).
Nucleic acid
Members of the family Sphaerolipoviridae have a linear double-stranded DNA genome of 28–31 kbp with the overall GC content of 67–68 % and inverted terminal repeats of about 300 bp in length with terminal proteins attached (Bamford et al., 2005, Porter et al., 2008, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016).
Proteins
12 genes have been shown to encode structural proteins (Table 2.Sphaerolipoviridae). The virion contains major and minor capsid proteins, internal membrane proteins and vertex complex proteins (Figure 1c.Sphaerolipoviridae) (Bamford et al., 2005, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016). The nomenclature of virion proteins (VPs) is the same for all sphaerolipoviruses. The most conserved structural proteins are MCPs (VP4 and VP7), the major membrane protein (protein VP12), and the putative packaging ATPase (putative protein 13), while the least conserved ones are vertex complex proteins (Demina et al., 2016, Santos-Pérez et al., 2019).
Table 2.Sphaerolipoviridae. HCIV-1 virion proteins.
| Protein | Calculated mass (kDa) | Location / Function |
| VP1 | 176.9 | Unknown |
| VP2 | 58.2 | Spike complex at the vertex (horn backbone) |
| VP3 | 36.7 | Spike complex at the vertex (horn) |
| VP4 | 26.0 | Large MCP |
| VP5 | 28.6 | Unknown |
| VP6 | 28.1 | Spike complex at the vertex (horn) |
| VP7 | 19.9 | Small MCP |
| VP9 | 16.3 | Penton protein at the vertex |
| VP10 | 17.7 | Major membrane-associated protein |
| VP12 | 9.8 | Major membrane-associated protein |
| VP13 | 8.8 | Unknown (putative packaging ATPase) |
| VP18 | 84.7 | Unknown |
Lipids
The virion contains lipids that form the internal membrane vesicle located underneath the icosahedral capsid. Lipids are selectively acquired from host cell membranes. The membrane vesicle is rich in virus-specific proteins. Major phospholipid species are phosphatidylglycerol, phosphatidylglycerophosphate methyl ester, and phosphatidylglycerosulfate (Bamford et al., 2005, Demina et al., 2016).
Carbohydrates
None reported
Genome organization and replication
Sphaerolipovirus genomes contain about 50 predicted open reading frames (ORFs) or genes, and a strong conservation of synteny is observed (Figure 2.Sphaerolioviridae) (Bamford et al., 2005, Porter et al., 2008, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016). The overall nucleotide identity between sphaerolipovirus genomes is 56–76 % (Demina et al., 2017). The replication mode is most probably protein-primed (Porter et al., 2008, Porter et al., 2013), but the genome does not include a gene for a canonical DNA polymerase. In the genome of SH1, putative protein-coding genes are organized in seven major transcripts, some of which overlap (Porter et al., 2008). Six SH1 transcripts, which encode structural genes, are synthesized early onwards starting at 1 h post infection and one, which encodes unknown proteins, is synthesized late in the infection cycle, at 5–6 h after infection. Based on the gene synteny (Figure 2.Sphaerolipoviridae), the transcriptional program seems to be the same for all sphaerolipoviruses.
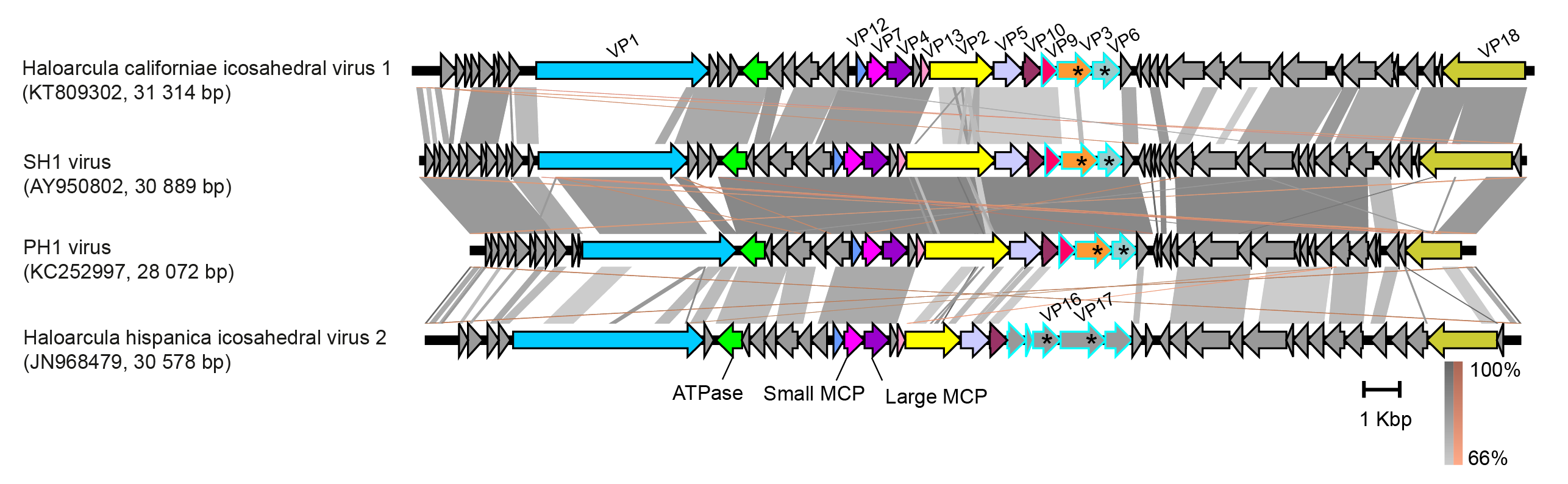 |
| Figure 2.Sphaerolipoviridae. Genomes of sphaerolipoviruses. ORFs/genes are shown as arrows with some protein products noted. Homologous genes encoding structural proteins are highlighted with the same colours. Putative spike protein genes are marked with asterisks and ORFs encoding for putative spike complex proteins are outlined with blue. Similarities between the genomes are shown as shadings of grey (direct) and brown (inverted). The figure was produced using Easyfig v.2.2.2 with the E-value threshold of 0.001(Sullivan et al., 2011). |
Biology
Sphaerolipoviruses were isolated from hypersaline environments in Europe, Asia, and Australia, such as solar salterns and salt lakes, and their host range is limited to a few (up to four) haloarchaeal strains belonging to the genera Haloarcula or Halorubrum (Porter et al., 2005, Atanasova et al., 2012, Porter et al., 2013, Atanasova et al., 2015). Transfection experiments with isolated virus genomes suggest the host range may also include Natrialba and Haloferax species (Porter and Dyall-Smith 2008, Porter et al., 2013). Sphaerolipoviruses bind to their hosts most probably by spike complexes at the virion vertices (Figure 1c.Sphaerolipoviridae). Adsorption rates are low, similar to those of many other haloarchaeal viruses (Kukkaro and Bamford 2009, Jaakkola et al., 2012, Demina et al., 2016). The infection cycle is lytic and lasts 6–12 hours, resulting in progeny virus production with a burst size of 50–200 viruses per cell (Porter et al., 2005, Jaakkola et al., 2012, Porter et al., 2013, Demina et al., 2016, Svirskaitė et al., 2016). Gene transcription is programmed and occurs during the early, middle and late stages of infection (Porter et al., 2008). Lipids are selectively obtained from the host cell membrane during virion assembly and form an inner membrane underneath the virion protein shell (Bamford et al., 2005, Jäälinoja et al., 2008, Gil-Carton et al., 2015, Demina et al., 2016, Santos-Pérez et al., 2019). Interactions between virus-encoded structural membrane proteins and the capsomers guide virion assembly (Jäälinoja et al., 2008, Gil-Carton et al., 2015). Sphaerolipoviruses have a gene for a packaging ATPase similar to that of the bacteriophage PRD1 (family Tectiviridae), which has been shown to package its linear dsDNA genome into a preformed capsid. Sphaerolipoviruses most probably package their DNA into preformed capsids, although this has not been experimentally shown (Porter et al., 2005, Jaakkola et al., 2012). Several putative proviral regions related to sphaerolipoviruses have been identified in the chromosomes of halophilic archaea (Demina et al., 2017).
Derivation of names
Sphaerolipoviridae: from the Latin sphaera, meaning “sphere” and the Greek lipos, meaning “fat” from which the term “lipid” is derived
Genus demarcation criteria
Since only one genus is currently recognized, the genus demarcation criteria have not been defined.
Relationships within the family
Members of the family share overall virion organization, sequence similarity and gene synteny.
Relationships with other taxa
Sphaerolipoviruses are characterized by a pseudo T = 28 dextro capsid lattice built of two vertical single jelly roll MCPs. The same lattice symmetry is also seen in Thermus bacteriophage P23-77 (Hukuchivirus P2377, family Matsushitaviridae) (Jaatinen et al., 2008). Structurally, sphaerolipovirus capsid organization resembles the lattice formed by the vertical double jelly roll MCPs, as seen for bacteriophage PRD1 (Alphatectivirus PRD1, family Tectiviridae, kingdom Bamfordvirae) (Benson et al., 1999). The family Sphaerolipoviridae together with the families Matsushitaviridae (species Hukuchivirus P2377 and Hukuchivirus IN93) and Simuloviridae (species Yingchengvirus SNJ1, Yingchengvirus NVIV1, and Yingchengvirus HJIV1) are assigned to the order Halopanivirales, class Laserviricetes, phylum Dividoviricota, kingdom Helvetiavirae. These three families (Sphaerolipoviridae, Matsushitaviridae and Simuloviridae) unite viruses that are structurally similar, all encoding two single jelly-roll MCPs and having an internal lipid membrane and DNA genome that can be either linear or circular. The kingdoms Helvetiavirae (tailless icosahedral viruses with two vertical single jelly-roll MCPs) and Bamfordvirae (tailless icosahedral viruses with a double jelly-roll MCP) form the realm Varidnaviria (Koonin et al., 2020).

