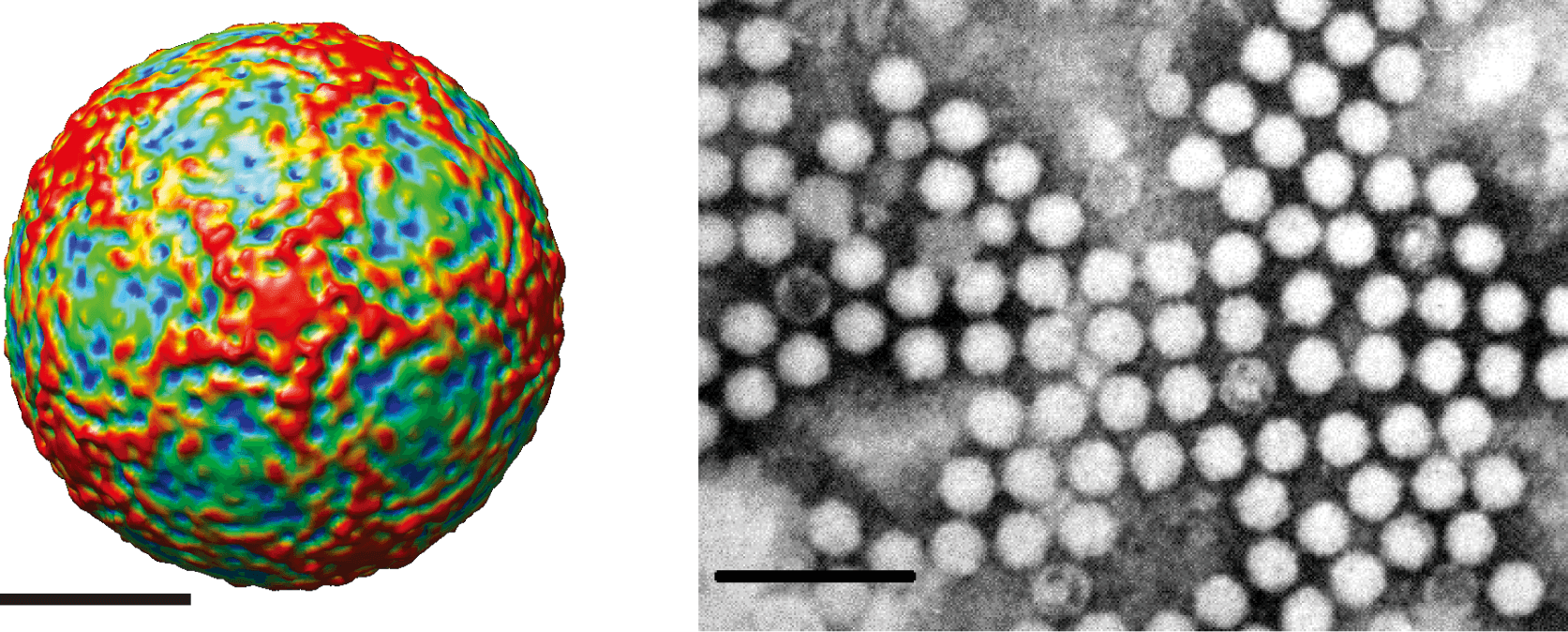Family: Iflaviridae
Yanping (Judy) Chen, Steven M. Valles, Andrew Firth, Joachim de Miranda, Eugene Ryabov, María G. Echeverría, Declan Schroeder, Eric Jan, Huoqing Zheng, and Rhys Parry
The citation for this ICTV Report chapter is the summary published as Valles et al., (2017):
ICTV Virus Taxonomy Profile: Iflaviridae, Journal of General Virology, 98, 527–528.
Corresponding author: Yanping Chen (judy.chen@usda.gov)
Edited by: Nick J. Knowles and Peter Simmonds
Posted: April 2017, updated May 2022, May 2025
PDF: ICTV_Iflaviridae.pdf (2017 version)
Summary
Iflaviridae is a family of small non-enveloped viruses with RNA genomes of approximately 9–11 kilobases in length encoding a single polyprotein (Table 1 Iflaviridae). All members infect arthropod hosts with the majority infecting insects. Beneficial and pest insects serve as hosts and infections can be symptomless (Nilaparvata lugens honeydew virus 1), cause developmental abnormalities (deformed wing virus, sacbrood virus), behavioral changes (deformed wing virus, slow bee paralysis virus) and premature mortality (deformed wing virus, slow bee paralysis virus, infectious flacherie virus, sacbrood virus) (van Oers 2010).
Table 1 Iflaviridae. Characteristics of members of the family Iflaviridae.
| Characteristic | Description |
| Example | infectious flacherie virus (AB000906), species Iflavirus flacherie |
| Virion | Non-enveloped, 22–30 nm diameter virions |
| Genome | 9–11 kb of positive-sense, non-segmented RNA |
| Replication | Cytoplasmic within viral replication complexes formed from a variety of host cellular membranes |
| Translation | Directly from genomic RNA containing an internal ribosomal entry site (IRES) |
| Host Range | Arthropoda |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Picornavirales; 16 species in the single genus Iflavirus |
Virion
Morphology
Virions are roughly spherical and exhibit icosahedral symmetry with a diameter of 22–30 nm. Virions have no envelope and no distinctive surface structures (Figure 1 Iflaviridae).
Physicochemical and physical properties
Virions have a buoyant density of between 1.29 and 1.38 g ml−1 in CsCl. Deformed wing virus and Varroa destructor virus 1 (both members of the species Iflavirus aladeformis) are unstable in isolation.
Nucleic acid
Virions contain one molecule of linear, positive-sense, single-stranded RNA approximately 9–11 kilobases in length containing a single large open reading frame (ORF) encoding a polyprotein of approximately 3,000 amino acids. A genome-linked virus protein, VPg, is covalently attached to the 5′-end of the genomic RNA and the 3′-terminus is polyadenylated. The non-coding regions (NCRs) flanking both ends of the ORF vary in size according to species.
Proteins
All proteins arise by proteolytic cleavage of the single polyprotein. Mature virions contain three major structural proteins (VP1, VP2 and VP3) generally between 28 and 44 kDa. The structural proteins are present in the N-terminal region of the polyprotein (Figure 2 Iflaviridae). A fourth smaller capsid protein (VP4) of around 4–12 kDa has been reported in some species and is located in the second position of the capsid precursor coding region. Minor quantities of the uncleaved precursors have been reported for some species. Non-structural proteins involved in replication and polyprotein processing are present in the C-terminal region of the polyprotein. The non-structural proteins include an RNA helicase (Gorbalenya et al., 1988), a 3C-like cysteine protease (Ye et al., 2012), and an RNA-directed RNA polymerase (Koonin and Dolja 1993), which are found in this order (Figure 2 Iflaviridae). Each of the non-structural proteins exhibits conserved, defining domains (van Oers 2010).
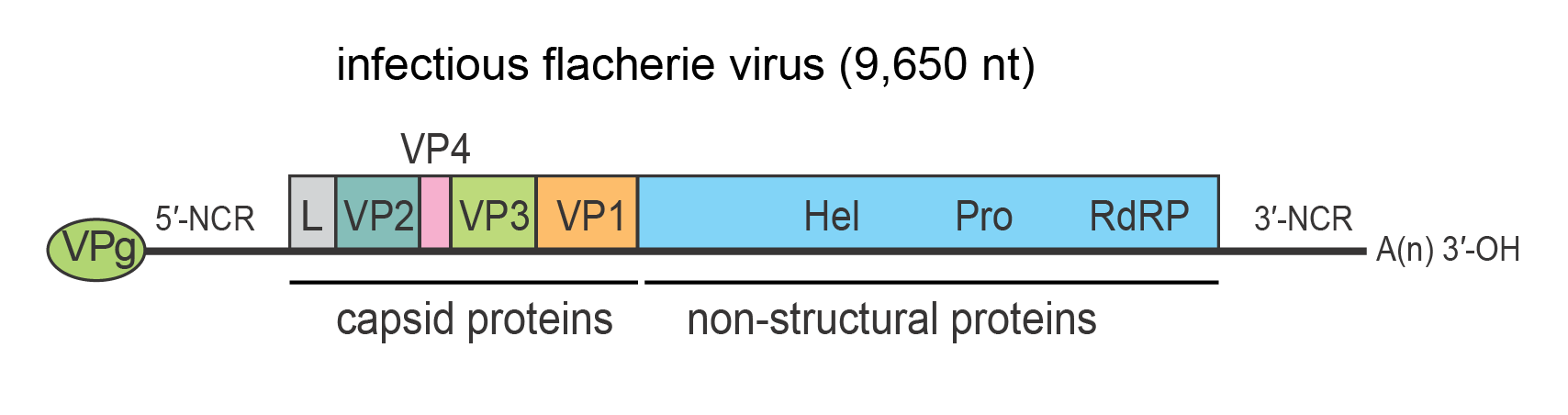 |
| Figure 2 Iflaviridae Genome structure of infectious flacherie virus. The genome encodes a single polyprotein that is auto-catalytically cleaved into three major structural proteins (VP1, VP2, VP3) and non-structural proteins used in replication. The 5′-end of the genome carries a covalently linked protein, VPg, which plays an important role in RNA replication and the 3′-terminus of the genome is polyadenylated. The structural proteins are encoded in the 5′-proximal region of the genome and the non-structural proteins in the 3′-proximal region. The capsid proteins, arranged in the order VP2-VP4-VP3-VP1, are preceded by a short leader protein (L). The approximate positions of the helicase (Hel), protease (Pro) and RNA-directed RNA polymerase (RdRP) domains are shown. |
Genome organization and replication
Iflaviruses possess single-stranded, positive-sense, non-segmented RNA genomes with a single ORF. The ORF is translated directly into a polyprotein that is subsequently processed to yield structural/capsid (N-terminal region) and non-structural (C-terminal region) proteins. Replication occurs in the host cell cytoplasm. The capsid proteins, arranged in the order of VP2-VP4-VP3-VP1, are often preceded by a short leader protein (L) of unknown function that is removed from VP2 before capsid assembly. VP4 is analogous to VP4 present in some dicistroviruses and in the case of infectious flacherie virus (IFV) is present as a minor structural component of the capsid. The non-structural proteins include an RNA helicase, a 3C-like cysteine protease, and an RNA-directed RNA polymerase (Figure 2 Iflaviridae). Translation initiation has been reported to be mediated by a 5′-NCR IRES in many iflaviruses, which is likely a common mechanism in the genus (Ongus et al., 2006, Lu et al., 2007). The viral RNA is infectious and serves as both genomic and viral mRNA. Mechanisms of polyprotein processing and the effects on host cell macromolecular synthesis during infection have not been well studied for the members of this family.
Biology
All member viruses have been isolated from arthropods. The host range of most members has not been examined. However, the honeybee iflaviruses including deformed wing virus, slow bee paralysis virus and sacbrood virus have been shown to infect other Apis species, as well as several Bombus species. Deformed wing virus also infects bee-parasitic mites (Varroa and Tropilaelaps spp.). Vertical and venereal transmission has been reported in the honey bee for deformed wing virus (de Miranda and Genersch 2010). The most common route of infection among the iflaviruses is through ingestion of virus-contaminated food sources. Trophallaxis in social insects facilitates intra-colonial virus dispersal. Deformed wing virus and slow bee paralysis virus can also be vectored to honeybees by parasitic mites (Varroa and Tropilaelaps genera). In addition to the gut, gonads, fat body, muscle, brain and glandular tissues also have been shown to be a target for several iflaviruses. Once the virus gains entry to the host cell, the infection process is rapid with progeny virus being produced in hours.
Antigenicity
Honeybee viruses in the genus are serologically distinct from each other. There are no known serological relationships between the other members of the genus.
Genus demarcation criteria
Sequence identity at the amino acid level between the capsid proteins of isolates and strains of a species is above 90%.
Derivation of names
Iflaviridae, Iflavirus: from the virus, infectious flacherie virus
Relationships within the family
Phylogenetic analysis of the complete translated genomes of iflaviruses show that a number of distinct clades exist (Figure 3 Iflaviridae). These will be likely separated taxonomically into different genera in the near future as more virus sequences become available. There does not appear to be an obvious host-based evolutionary relationship because each clade is composed of viruses infecting hosts from different insect orders.
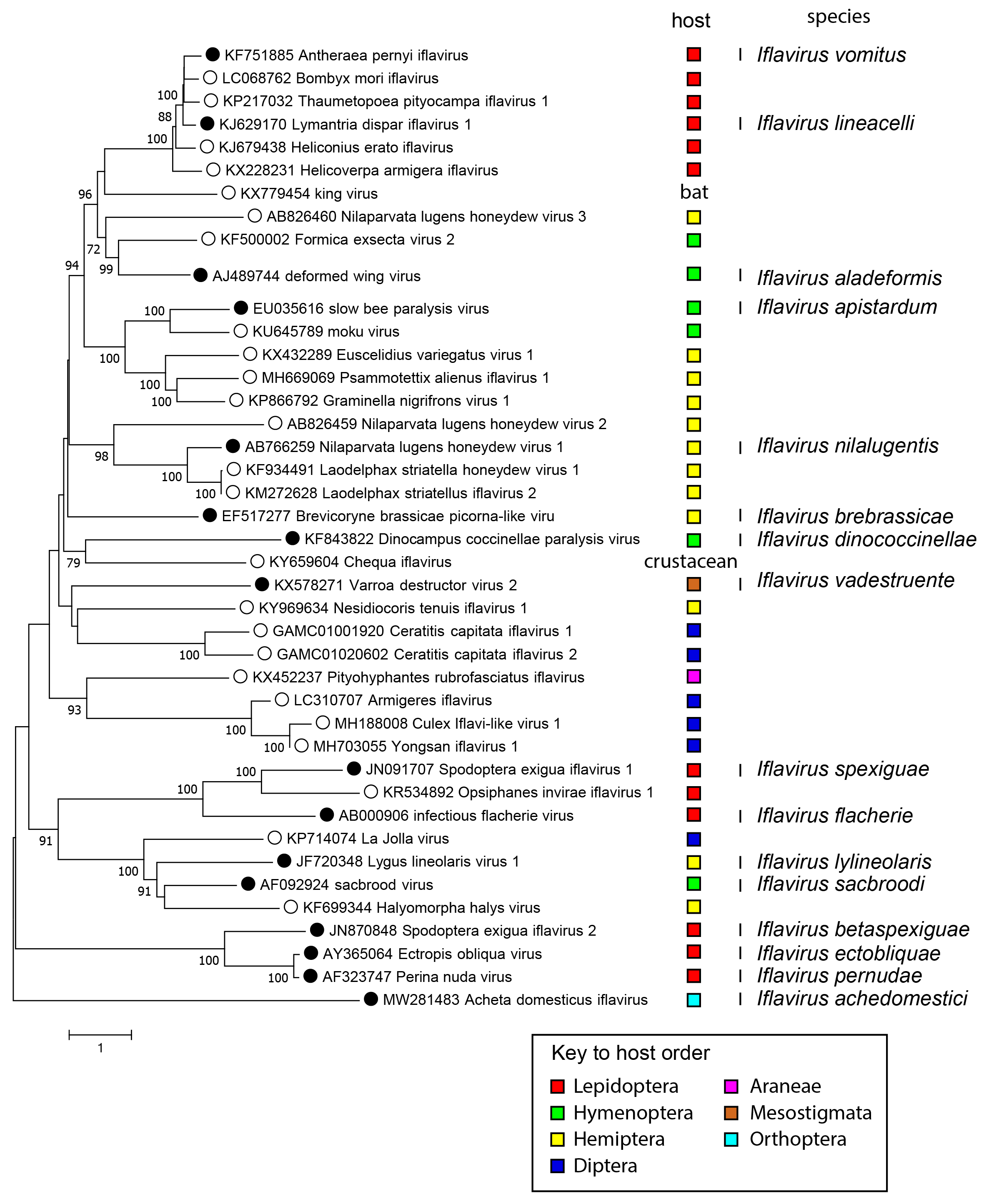 |
| Figure 3 Iflaviridae Mid-point rooted phylogenetic tree. Complete polyprotein sequences were aligned with MUSCLE (Edgar 2004), and a phylogenetic tree produced based on amino acid distances using MEGA7 (Kumar et al., 2016). Numbers at nodes indicate bootstrap support where this was >70%. The order of the presumed host is indicated by coloured squares. Viruses that have been classified into one of the 16 Iflavirus species are indicated with a solid dot along with the species name in a separate column. |
Relationships with other taxa
The family Iflaviridae is a member of the order Picornavirales. Iflaviruses share properties with other members of the order (Dicistroviridae, Marnaviridae, Picornaviridae, and Secoviridae), including three replication proteins (helicase, protease, RNA-directed RNA polymerase), non-enveloped icosahedral virions of approximately 30 nm diameter, presence of a small VPg protein at the 5′-terminus, and a polyadenylated 3′-terminus.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Bombyx mori iflavirus | LC068762 | BMIV |
| Ceratitis capitata iflavirus 1 | GAMC01001920 | CcIV1 |
| Ceratitis capitata iflavirus 2 | GAMC01020602 | CcIV2 |
| Formica exsecta virus 2 | KF500002 | FeV2 |
| Graminella nigrifrons virus 1 | KP866792 | GnV1 |
| Halyomorpha halys virus | KF699344 | HhV |
| Heliconius erato iflavirus | KJ679438 | HeIV |
| king virus | KX779454 | KgV |
| kinkell virus | KU754510 | KkV |
| La Jolla virus | KP714074 | LJV |
| Laodelphax striatella honeydew virus 1 | KF934491 | LsHV1 |
| Laodelphax striatellus iflavirus 2 | KM272628 | LsIV2 |
| moku virus | NC031338 | MV |
| Nilaparvata lugens honeydew virus 2 | AB826459 | NlHV2 |
| Nilaparvata lugens honeydew virus 3 | AB826460 | NlHV3 |
| Osiphanes invirae iflavirus 1 | KR534892 | OiIV1 |
| Thaumetopoea pityocampa iflavirus 1 | KP217032 | TpIV1 |
| Chequa iflavirus | KY659604 | CIV |
| Culex Iflavi-like virus 1 | MH188008 | CILV1 |
| Euscelidius variegatus virus 1 | KX432289 | EVV1 |
| Pityohyphantes rubrofasciatus iflavirus | KX452237 | PRIV |
| Yongsan iflavirus 1 | MH703055 | YIV1 |
| Nesidiocoris tenuis iflavirus 1 | KY969634 | NTIV1 |
Virus names and virus abbreviations are not official ICTV designations.